2J82
 
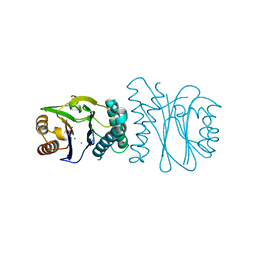 | | Structural analysis of the PP2C Family Phosphatase tPphA from Thermosynechococcus elongatus | | Descriptor: | CALCIUM ION, MAGNESIUM ION, PROTEIN SERINE-THREONINE PHOSPHATASE | | Authors: | Schlicker, C, Kloft, N, Forchhammer, K, Becker, S. | | Deposit date: | 2006-10-18 | | Release date: | 2007-11-06 | | Last modified: | 2017-10-18 | | Method: | X-RAY DIFFRACTION (1.28 Å) | | Cite: | Structural Analysis of the Pp2C Phosphatase Tppha from Thermosynechococcus Elongatus: A Flexible Flap Subdomain Controls Access to the Catalytic Site.
J.Mol.Biol., 376, 2008
|
|
2J86
 
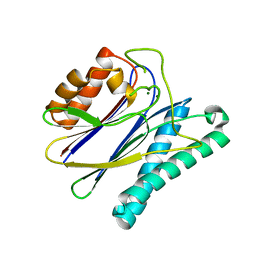 | | Structural analysis of the PP2C Family Phosphatase tPphA of Thermosynechococcus elongatus | | Descriptor: | MAGNESIUM ION, PROTEIN SERINE-THREONINE PHOSPHATASE | | Authors: | Schlicker, C, Kloft, N, Forchhammer, K, Becker, S. | | Deposit date: | 2006-10-19 | | Release date: | 2007-11-06 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (3.05 Å) | | Cite: | Structural Analysis of the Pp2C Phosphatase Tppha from Thermosynechococcus Elongatus: A Flexible Flap Subdomain Controls Access to the Catalytic Site.
J.Mol.Biol., 376, 2008
|
|
2W3N
 
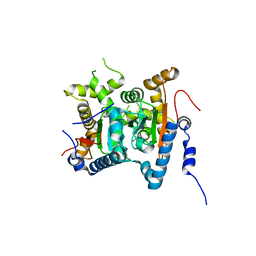 | | Structure and inhibition of the CO2-sensing carbonic anhydrase Can2 from the pathogenic fungus Cryptococcus neoformans | | Descriptor: | ACETATE ION, CARBONIC ANHYDRASE 2, GLYCEROL, ... | | Authors: | Schlicker, C, Hall, R.A, Vullo, D, Middelhaufe, S, Gertz, M, Supuran, C.T, Muehlschlegel, F.A, Steegborn, C. | | Deposit date: | 2008-11-13 | | Release date: | 2008-12-30 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structure and inhibition of the CO2-sensing carbonic anhydrase Can2 from the pathogenic fungus Cryptococcus neoformans.
J. Mol. Biol., 385, 2009
|
|
2W3Q
 
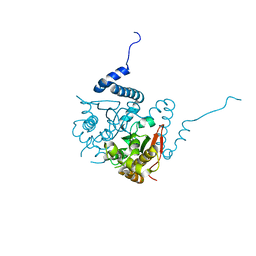 | | Structure and inhibition of the CO2-sensing carbonic anhydrase Can2 from the pathogenic fungus Cryptococcus neoformans | | Descriptor: | CARBONIC ANHYDRASE 2, CHLORIDE ION, ZINC ION | | Authors: | Schlicker, C, Hall, R.A, Vullo, D, Middelhaufe, S, Gertz, M, Supuran, C.T, Muehlschlegel, F.A, Steegborn, C. | | Deposit date: | 2008-11-14 | | Release date: | 2008-12-30 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.34 Å) | | Cite: | Structure and Inhibition of the Co(2)-Sensing Carbonic Anhydrase Can2 from the Pathogenic Fungus Cryptococcus Neoformans.
J.Mol.Biol., 385, 2009
|
|
2CM5
 
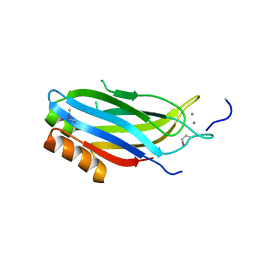 | | crystal structure of the C2B domain of rabphilin | | Descriptor: | CALCIUM ION, RABPHILIN-3A | | Authors: | Schlicker, C, Montaville, P, Sheldrick, G.M, Becker, S. | | Deposit date: | 2006-05-04 | | Release date: | 2006-12-04 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.28 Å) | | Cite: | The C2A-C2B Linker Defines the High Affinity Ca2+ Binding Mode of Rabphilin-3A.
J.Biol.Chem., 282, 2007
|
|
2CM6
 
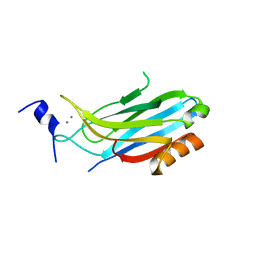 | | crystal structure of the C2B domain of rabphilin3A | | Descriptor: | CALCIUM ION, PHOSPHATE ION, RABPHILIN-3A | | Authors: | Schlicker, C, Montaville, P, Sheldrick, G.M, Becker, S. | | Deposit date: | 2006-05-04 | | Release date: | 2006-12-04 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | The C2A-C2B Linker Defines the High Affinity Ca2+ Binding Mode of Rabphilin-3A.
J.Biol.Chem., 282, 2007
|
|
2XSS
 
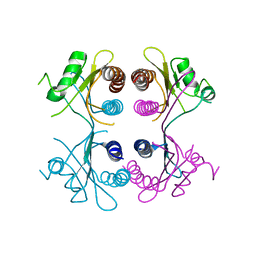 | |
2XZV
 
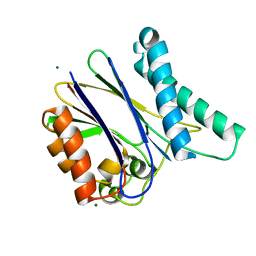 | |
2Y09
 
 | |
4TR4
 
 | |
4TR3
 
 | |
6XTU
 
 | | FULL-LENGTH LTTR LYSG FROM CORYNEBACTERIUM GLUTAMICUM | | Descriptor: | Lysine export transcriptional regulatory protein LysG | | Authors: | Hofmann, E, Syberg, F, Schlicker, C, Eggeling, L, Schendzielorz, G. | | Deposit date: | 2020-01-16 | | Release date: | 2020-10-07 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.52 Å) | | Cite: | Engineering and application of a biosensor with focused ligand specificity.
Nat Commun, 11, 2020
|
|
6XTV
 
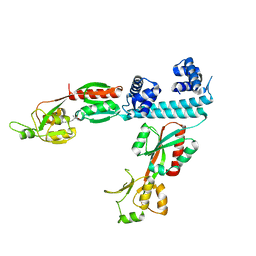 | | FULL-LENGTH LTTR LYSG FROM CORYNEBACTERIUM GLUTAMICUM WITH BOUND EFFECTOR ARG | | Descriptor: | ARGININE, Lysine export transcriptional regulatory protein LysG | | Authors: | Hofmann, E, Syberg, F, Schlicker, C, Eggeling, L, Schendzielorz, G. | | Deposit date: | 2020-01-16 | | Release date: | 2020-10-07 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Engineering and application of a biosensor with focused ligand specificity.
Nat Commun, 11, 2020
|
|
2YJ4
 
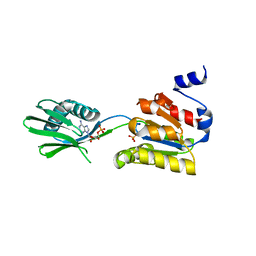 | | Conformational changes in the catalytic domain of the CPx-ATPase CopB- B upon nucleotide binding | | Descriptor: | ADENOSINE MONOPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, COPPER-TRANSPORTING ATPASE, ... | | Authors: | Voellmecke, C, Schlicker, C, Luebben, M, Hofmann, E. | | Deposit date: | 2011-05-18 | | Release date: | 2012-05-30 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Conformational Changes in the Catalytic Domain of the Cpx-ATPase Copb-B Upon Nucleotide Binding
To be Published
|
|
2YJ3
 
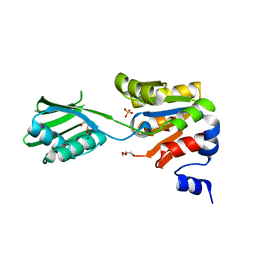 | | Conformational changes in the catalytic domain of the CPx-ATPase CopB- B upon nucleotide binding | | Descriptor: | COPPER-TRANSPORTING ATPASE, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Voellmecke, C, Schlicker, C, Luebben, M, Hofmann, E. | | Deposit date: | 2011-05-18 | | Release date: | 2012-05-30 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Conformational Changes in the Catalytic Domain of the Cpx-ATPase Copb-B Upon Nucleotide Binding
To be Published
|
|
2YJ5
 
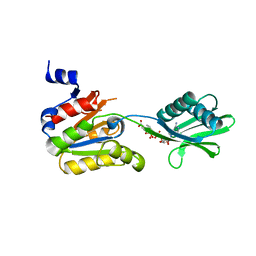 | |
2YJ6
 
 | |
4HDA
 
 | |
4HD8
 
 | | Crystal structure of human Sirt3 in complex with Fluor-de-Lys peptide and piceatannol | | Descriptor: | Fluor-de-Lys peptide, ISOPROPYL ALCOHOL, NAD-dependent protein deacetylase sirtuin-3, ... | | Authors: | Nguyen, G.T.T, Gertz, M, Steegborn, C. | | Deposit date: | 2012-10-02 | | Release date: | 2012-12-05 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | A molecular mechanism for direct sirtuin activation by resveratrol.
Plos One, 7, 2012
|
|
1O9W
 
 | | F17-aG lectin domain from Escherichia coli in complex with N-acetyl-glucosamine | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, F17A-G FIMBRIAL ADHESIN | | Authors: | Buts, L, De Genst, E, Loris, R, Oscarson, S, Lahmann, M, Messens, J, Brosens, E, Wyns, L, Bouckaert, J, De Greve, H. | | Deposit date: | 2002-12-20 | | Release date: | 2003-05-29 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | The Fimbrial Adhesin F17-G of Enterotoxigenic Escherichia Coli Has an Immunoglobulin-Like Lectin Domain that Binds N-Acetylglucosamine
Mol.Microbiol., 49, 2003
|
|
1O9V
 
 | | F17-aG lectin domain from Escherichia coli in complex with a selenium carbohydrate derivative | | Descriptor: | F17-AG LECTIN DOMAIN, methyl 2-acetamido-2-deoxy-1-seleno-beta-D-glucopyranoside | | Authors: | Buts, L, De Genst, E, Loris, R, Oscarson, S, Lahmann, M, Messens, J, Brosens, E, Wyns, L, Bouckaert, J, De Greve, H. | | Deposit date: | 2002-12-20 | | Release date: | 2003-05-29 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | The Fimbrial Adhesin F17-G of Enterotoxigenic Escherichia Coli Has an Immunoglobulin-Like Lectin Domain that Binds N-Acetylglucosamine
Mol.Microbiol., 49, 2003
|
|
1O9Z
 
 | | F17-aG lectin domain from Escherichia coli (ligand free) | | Descriptor: | F17-AG LECTIN DOMAIN | | Authors: | Buts, L, De Genst, E, Loris, R, Oscarson, S, Lahmann, M, Messens, J, Brosens, E, Wyns, L, Bouckaert, J, De Greve, H. | | Deposit date: | 2002-12-23 | | Release date: | 2003-05-29 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | The Fimbrial Adhesin F17-G of Enterotoxigenic Escherichia Coli Has an Immunoglobulin-Like Lectin Domain that Binds N-Acetylglucosamine
Mol.Microbiol., 49, 2003
|
|
