8TCJ
 
 | |
8TCK
 
 | |
8TCL
 
 | |
8TCM
 
 | | Crystal Structure of modified HIV reverse transcriptase p51 domain (FPC1) with picric acid and Xanthene-1,3,6,8-tetrol bound | | Descriptor: | 9H-xanthene-1,3,6,8-tetrol, PICRIC ACID, p51 subunit | | Authors: | Pedersen, L.C, London, R.E. | | Deposit date: | 2023-07-02 | | Release date: | 2024-07-17 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Crystal Structure of modified HIV reverse transcriptase p51 domain (FPC1) with picric acid and xanthene bound
To Be Published
|
|
4Q5R
 
 | |
4Q5Q
 
 | |
4Q5N
 
 | |
4Q5F
 
 | |
1ON6
 
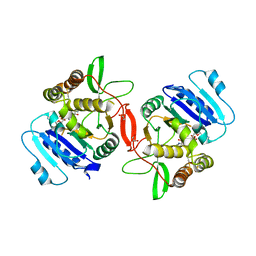 | | Crystal structure of mouse alpha-1,4-N-acetylhexosaminotransferase (EXTL2) in complex with UDPGlcNAc | | Descriptor: | 1,2-ETHANEDIOL, Alpha-1,4-N-acetylhexosaminyltransferase EXTL2, MANGANESE (II) ION, ... | | Authors: | Pedersen, L.C, Dong, J, Taniguchi, F, Kitagawa, H, Krahn, J.M, Pedersen, L.G, Sugahara, K, Negishi, M. | | Deposit date: | 2003-02-27 | | Release date: | 2003-04-22 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structure of an alpha-1,4-N-acetylhexosaminyltransferase (EXTL2), a member of the exostosin gene family involved in heparan sulfate biosynthesis
J.Biol.Chem., 278, 2003
|
|
1OMZ
 
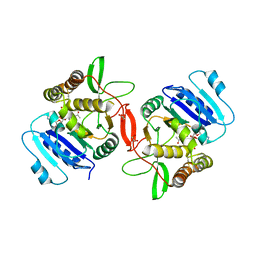 | | crystal structure of mouse alpha-1,4-N-acetylhexosaminyltransferase (EXTL2) in complex with UDPGalNAc | | Descriptor: | 1,2-ETHANEDIOL, Alpha-1,4-N-acetylhexosaminyltransferase EXTL2, MANGANESE (II) ION, ... | | Authors: | Pedersen, L.C, Dong, J, Taniguchi, F, Kitagawa, H, Krahn, J.M, Pedersen, L.G, Sugahara, K, Negishi, M. | | Deposit date: | 2003-02-26 | | Release date: | 2003-04-22 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of an alpha-1,4-N-acetylhexosaminyltransferase (EXTL2),
a member of the exostosin gene family involved in heparan sulfate biosynthesis
J.Biol.Chem., 278, 2003
|
|
1OMX
 
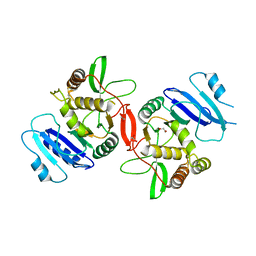 | | Crystal structure of mouse alpha-1,4-N-acetylhexosaminyltransferase (EXTL2) | | Descriptor: | 1,2-ETHANEDIOL, Alpha-1,4-N-acetylhexosaminyltransferase EXTL2 | | Authors: | Pedersen, L.C, Dong, J, Taniguchi, F, Kitagawa, H, Krahn, J.M, Pedersen, L.G, Sugahara, K, Negishi, M. | | Deposit date: | 2003-02-26 | | Release date: | 2003-04-22 | | Last modified: | 2017-10-11 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of an alpha-1,4-N-acetylhexosaminyltransferase (EXTL2), a member of the exostosin gene family involved in heparan sulfate biosynthesis
J.Biol.Chem., 278, 2003
|
|
1ON8
 
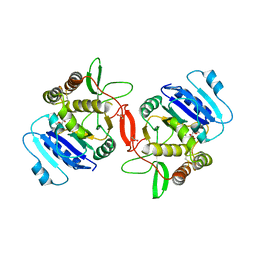 | | Crystal structure of mouse alpha-1,4-N-acetylhexosaminyltransferase (EXTL2) with UDP and GlcUAb(1-3)Galb(1-O)-naphthalenelmethanol an acceptor substrate analog | | Descriptor: | 1,2-ETHANEDIOL, Alpha-1,4-N-acetylhexosaminyltransferase EXTL2, MANGANESE (II) ION, ... | | Authors: | Pedersen, L.C, Dong, J, Taniguchi, F, Kitagawa, H, Krahn, J.M, Pedersen, L.G, Sugahara, K, Negishi, M. | | Deposit date: | 2003-02-27 | | Release date: | 2003-04-22 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of an alpha-1,4-N-acetylhexosaminyltransferase (EXTL2), a member of the exostosin gene family involved in heparan sulfate biosynthesis
J.Biol.Chem., 278, 2003
|
|
6MW7
 
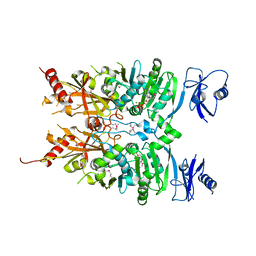 | | Crystal structure of ATPase module of SMCHD1 bound to ATP | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, SODIUM ION, ... | | Authors: | Pedersen, L.C, Inoue, K, Kim, S, Perera, L, Shaw, N.D. | | Deposit date: | 2018-10-29 | | Release date: | 2019-09-11 | | Last modified: | 2019-12-18 | | Method: | X-RAY DIFFRACTION (2.194 Å) | | Cite: | A ubiquitin-like domain is required for stabilizing the N-terminal ATPase module of human SMCHD1.
Commun Biol, 2, 2019
|
|
1KWS
 
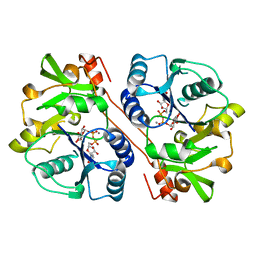 | | CRYSTAL STRUCTURE OF BETA1,3-GLUCURONYLTRANSFERASE I IN COMPLEX WITH THE ACTIVE UDP-GLCUA DONOR | | Descriptor: | BETA-1,3-GLUCURONYLTRANSFERASE 3, MANGANESE (II) ION, URIDINE-5'-DIPHOSPHATE-GLUCURONIC ACID | | Authors: | Pedersen, L.C, Darden, T.A, Negishi, M. | | Deposit date: | 2002-01-30 | | Release date: | 2002-06-19 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of beta 1,3-glucuronyltransferase I in complex with active donor substrate UDP-GlcUA.
J.Biol.Chem., 277, 2002
|
|
1HY3
 
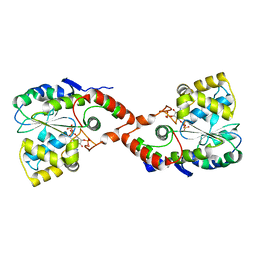 | | CRYSTAL STRUCTURE OF HUMAN ESTROGEN SULFOTRANSFERASE V269E MUTANT IN THE PRESENCE OF PAPS | | Descriptor: | 3'-PHOSPHATE-ADENOSINE-5'-PHOSPHATE SULFATE, ESTROGEN SULFOTRANSFERASE | | Authors: | Pedersen, L.C, Petrochenko, E.V, Shevtsov, S, Negishi, M. | | Deposit date: | 2001-01-17 | | Release date: | 2002-06-19 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of the human estrogen sulfotransferase-PAPS complex: evidence for catalytic role of Ser137 in the sulfuryl transfer reaction.
J.Biol.Chem., 277, 2002
|
|
1KNY
 
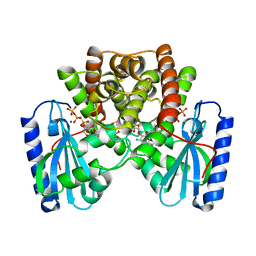 | | KANAMYCIN NUCLEOTIDYLTRANSFERASE | | Descriptor: | DIPHOSPHOMETHYLPHOSPHONIC ACID ADENOSYL ESTER, KANAMYCIN A, KANAMYCIN NUCLEOTIDYLTRANSFERASE, ... | | Authors: | Pedersen, L.C, Benning, M.M, Holden, H.M. | | Deposit date: | 1995-07-07 | | Release date: | 1996-08-17 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural investigation of the antibiotic and ATP-binding sites in kanamycin nucleotidyltransferase.
Biochemistry, 34, 1995
|
|
1EFH
 
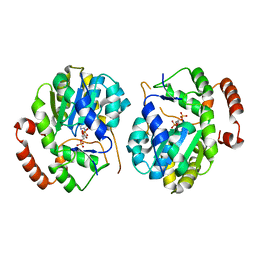 | |
1FGG
 
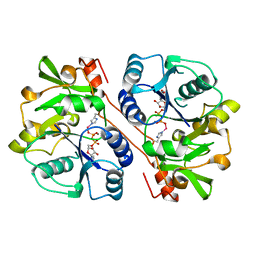 | | CRYSTAL STRUCTURE OF 1,3-GLUCURONYLTRANSFERASE I (GLCAT-I) COMPLEXED WITH GAL-GAL-XYL, UDP, AND MN2+ | | Descriptor: | GLUCURONYLTRANSFERASE I, MANGANESE (II) ION, UNKNOWN ATOM OR ION, ... | | Authors: | Pedersen, L.C, Tsuchida, K, Kitagawa, H, Sugahara, K, Darden, T.A. | | Deposit date: | 2000-07-28 | | Release date: | 2001-01-31 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Heparan/chondroitin sulfate biosynthesis. Structure and mechanism of human glucuronyltransferase I.
J.Biol.Chem., 275, 2000
|
|
5W4G
 
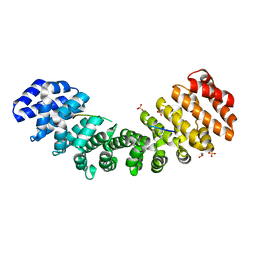 | |
8G4P
 
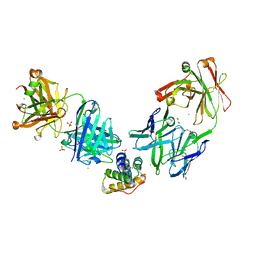 | | Crystal structure of the peanut allergen Ara h 2 bound by two neutralizing antibodies 13T1 and 13T5 | | Descriptor: | 1,2-ETHANEDIOL, 13T1 Fab light chain, 13T5 Fab heavy chain, ... | | Authors: | Pedersen, L.C, Mueller, G.A, Min, J. | | Deposit date: | 2023-02-10 | | Release date: | 2023-12-20 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Design of an Ara h 2 hypoallergen from conformational epitopes.
Clin Exp Allergy, 54, 2024
|
|
5T03
 
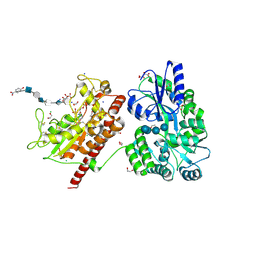 | | Crystal structure of heparan sulfate 6-O-sulfotransferase with bound PAP and glucuronic acid containing hexasaccharide substrate | | Descriptor: | 1,2-ETHANEDIOL, 2-deoxy-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-beta-D-glucopyranuronic acid-(1-4)-2-deoxy-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-beta-D-glucopyranuronic acid-(1-4)-2-deoxy-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-beta-D-glucopyranuronic acid, ADENOSINE-3'-5'-DIPHOSPHATE, ... | | Authors: | Pedersen, L.C, Moon, A.F, Krahn, J.M, Liu, J. | | Deposit date: | 2016-08-15 | | Release date: | 2017-02-01 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure Based Substrate Specificity Analysis of Heparan Sulfate 6-O-Sulfotransferases.
ACS Chem. Biol., 12, 2017
|
|
5T0A
 
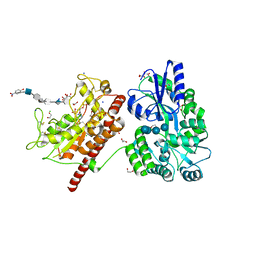 | | Crystal Structure of Heparan Sulfate 6-O-Sulfotransferase with bound PAP and heptasaccharide substrate | | Descriptor: | 1,2-ETHANEDIOL, 2-deoxy-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-beta-D-glucopyranuronic acid-(1-4)-2-deoxy-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-beta-D-glucopyranuronic acid, ADENOSINE-3'-5'-DIPHOSPHATE, ... | | Authors: | Pedersen, L.C, Moon, A.F, Krahn, J.M, Liu, J. | | Deposit date: | 2016-08-15 | | Release date: | 2017-02-01 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structure Based Substrate Specificity Analysis of Heparan Sulfate 6-O-Sulfotransferases.
ACS Chem. Biol., 12, 2017
|
|
5T05
 
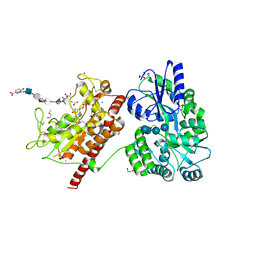 | | Crystal structure of heparan sulfate 6-O-sulfotransferase with bound PAP and IdoA2S containing hexasaccharide substrate | | Descriptor: | 1,2-ETHANEDIOL, 2-deoxy-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-beta-D-glucopyranuronic acid-(1-4)-2-deoxy-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-beta-D-glucopyranuronic acid, ADENOSINE-3'-5'-DIPHOSPHATE, ... | | Authors: | Pedersen, L.C, Moon, A.F, krahn, J.M, Liu, J. | | Deposit date: | 2016-08-15 | | Release date: | 2017-02-01 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.952 Å) | | Cite: | Structure Based Substrate Specificity Analysis of Heparan Sulfate 6-O-Sulfotransferases.
ACS Chem. Biol., 12, 2017
|
|
6D7N
 
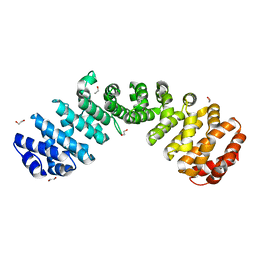 | |
6D7M
 
 | | Crystal structure of the W184R/W231R Importin alpha mutant | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Peroxidase,Importin subunit alpha-1, ... | | Authors: | Pedersen, L.C, London, R.E, Gabel, S.A. | | Deposit date: | 2018-04-25 | | Release date: | 2019-03-06 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.187 Å) | | Cite: | Variations in nuclear localization strategies among pol X family enzymes.
Traffic, 2018
|
|
