7UOW
 
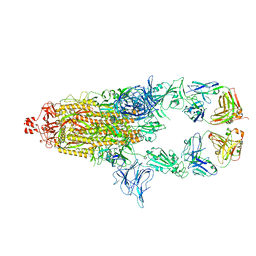 | | SARS-Cov2 S protein structure in complex with neutralizing monoclonal antibody 034_32 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Monoclonal antibody 034_32 heavy chain, ... | | Authors: | Patel, A, Ortlund, E. | | Deposit date: | 2022-04-14 | | Release date: | 2023-04-19 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (4.4 Å) | | Cite: | Molecular basis of SARS-CoV-2 Omicron variant evasion from shared neutralizing antibody response.
Structure, 31, 2023
|
|
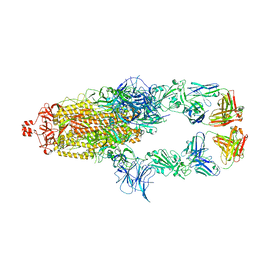
7U0Q
 
 | |
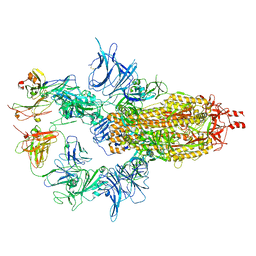
7U0X
 
 | |
6HPR
 
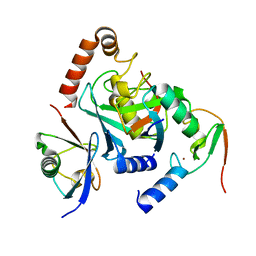 | |
3EG6
 
 | |
3M4A
 
 | | Crystal structure of a bacterial topoisomerase IB in complex with DNA reveals a secondary DNA binding site | | Descriptor: | ACETIC ACID, DNA (5'-D(*GP*AP*AP*TP*AP*AP*GP*GP*GP*CP*GP*C)-3'), DNA (5'-D(*GP*CP*GP*CP*CP*CP*TP*TP*AP*TP*TP*C)-3'), ... | | Authors: | Patel, A, Yakovleva, L, Shuman, S, Mondragon, A. | | Deposit date: | 2010-03-10 | | Release date: | 2010-07-14 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Crystal structure of a bacterial topoisomerase IB in complex with DNA reveals a secondary DNA binding site.
Structure, 18, 2010
|
|
8GDR
 
 | |
2F4Q
 
 | |
5EH2
 
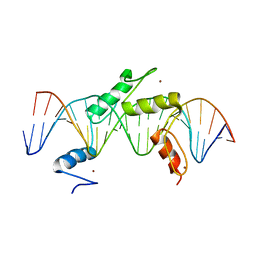 | | Human PRDM9 allele-A ZnF Domain with Associated Recombination Hotspot DNA Sequence III | | Descriptor: | DNA (5'-D(*AP*CP*AP*CP*GP*TP*GP*GP*CP*TP*AP*GP*GP*GP*AP*GP*GP*CP*CP*TP*C)-3'), DNA (5'-D(*TP*GP*AP*GP*GP*CP*CP*TP*CP*CP*CP*TP*AP*GP*CP*CP*AP*CP*GP*TP*G)-3'), Histone-lysine N-methyltransferase PRDM9, ... | | Authors: | Patel, A, Horton, J.R, Wilson, G.G, Zhang, X, Cheng, X. | | Deposit date: | 2015-10-27 | | Release date: | 2016-02-17 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structural basis for human PRDM9 action at recombination hot spots.
Genes Dev., 30, 2016
|
|
5EI9
 
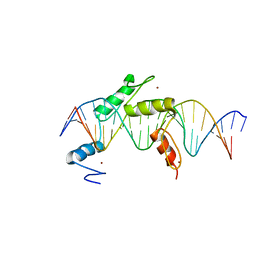 | | Human PRDM9 allele-A ZnF Domain with Associated Recombination Hotspot DNA Sequence I | | Descriptor: | DNA (5'-D(*AP*TP*CP*CP*AP*CP*GP*TP*GP*GP*CP*TP*AP*GP*GP*GP*AP*GP*GP*CP*C)-3'), DNA (5'-D(*TP*GP*GP*CP*CP*TP*CP*CP*CP*TP*AP*GP*CP*CP*AP*CP*GP*TP*GP*GP*A)-3'), Histone-lysine N-methyltransferase PRDM9, ... | | Authors: | Patel, A, Horton, J.R, Wilson, G.G, Zhang, X, Cheng, X. | | Deposit date: | 2015-10-29 | | Release date: | 2016-02-17 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.921 Å) | | Cite: | Structural basis for human PRDM9 action at recombination hot spots.
Genes Dev., 30, 2016
|
|
5EGB
 
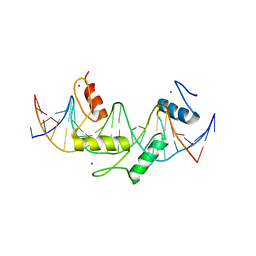 | | Human PRDM9 allele-A ZnF Domain with Associated Recombination Hotspot DNA Sequence II | | Descriptor: | DNA (5'-D(*AP*CP*CP*AP*CP*GP*TP*GP*GP*CP*TP*AP*GP*GP*GP*AP*GP*GP*CP*CP*T)-3'), DNA (5'-D(*TP*AP*GP*GP*CP*CP*TP*CP*CP*CP*TP*AP*GP*CP*CP*AP*CP*GP*TP*GP*G)-3'), Histone-lysine N-methyltransferase PRDM9, ... | | Authors: | Patel, A, Horton, J.R, Wilson, G.G, Zhang, X, Cheng, X. | | Deposit date: | 2015-10-26 | | Release date: | 2016-02-17 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.977 Å) | | Cite: | Structural basis for human PRDM9 action at recombination hot spots.
Genes Dev., 30, 2016
|
|
5V3J
 
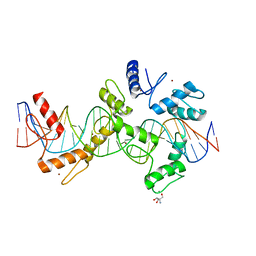 | | mouseZFP568-ZnF1-10 in complex with DNA | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, DNA (26-MER), MAGNESIUM ION, ... | | Authors: | Patel, A, Cheng, X. | | Deposit date: | 2017-03-07 | | Release date: | 2018-03-07 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.064 Å) | | Cite: | DNA Conformation Induces Adaptable Binding by Tandem Zinc Finger Proteins.
Cell, 173, 2018
|
|
5V3M
 
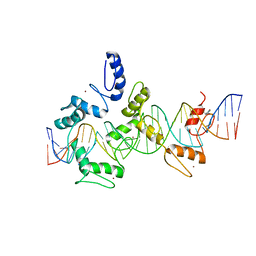 | | mouseZFP568-ZnF1-11 in complex with DNA | | Descriptor: | DNA (28-MER), ZINC ION, Zinc finger protein 568 | | Authors: | Patel, A, Cheng, X. | | Deposit date: | 2017-03-07 | | Release date: | 2018-03-07 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.091 Å) | | Cite: | DNA Conformation Induces Adaptable Binding by Tandem Zinc Finger Proteins.
Cell, 173, 2018
|
|
7MIA
 
 | | Maize rayado fino virus protease | | Descriptor: | RNA replication protein | | Authors: | Patel, A, Mark, B.L. | | Deposit date: | 2021-04-16 | | Release date: | 2021-07-21 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The endopeptidase of the maize-affecting Marafivirus type member maize rayado fino virus doubles as a deubiquitinase.
J.Biol.Chem., 297, 2021
|
|
7MIC
 
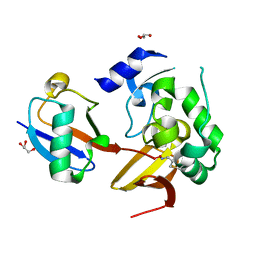 | | Maize rayado fino virus protease in complex with Ubiquitin | | Descriptor: | 3-AMINOPROPANE, GLYCEROL, RNA replication protein, ... | | Authors: | Patel, A, Mark, B.L. | | Deposit date: | 2021-04-16 | | Release date: | 2021-07-21 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | The endopeptidase of the maize-affecting Marafivirus type member maize rayado fino virus doubles as a deubiquitinase.
J.Biol.Chem., 297, 2021
|
|
5V3G
 
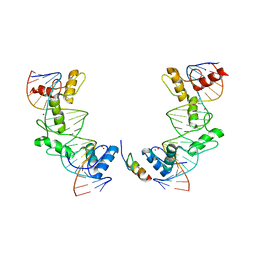 | | PRDM9-allele-C ZnF8-13 | | Descriptor: | DNA (5'-D(*AP*GP*GP*GP*CP*AP*AP*CP*GP*CP*TP*CP*AP*CP*TP*GP*GP*GP*GP*TP*C)-3'), DNA (5'-D(*TP*GP*AP*CP*CP*CP*CP*AP*GP*TP*GP*AP*GP*CP*GP*TP*TP*GP*CP*CP*C)-3'), PR domain zinc finger protein 9, ... | | Authors: | Patel, A, Cheng, X. | | Deposit date: | 2017-03-07 | | Release date: | 2017-08-23 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.416 Å) | | Cite: | Structural basis of human PR/SET domain 9 (PRDM9) allele C-specific recognition of its cognate DNA sequence.
J. Biol. Chem., 292, 2017
|
|
5WJQ
 
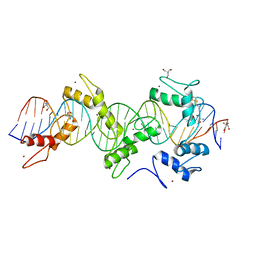 | | mouseZFP568-ZnF2-11 in complex with DNA | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, DNA (28-MER), ZINC ION, ... | | Authors: | Patel, A, Cheng, X. | | Deposit date: | 2017-07-24 | | Release date: | 2018-03-07 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.794 Å) | | Cite: | DNA Conformation Induces Adaptable Binding by Tandem Zinc Finger Proteins.
Cell, 173, 2018
|
|
7ZJS
 
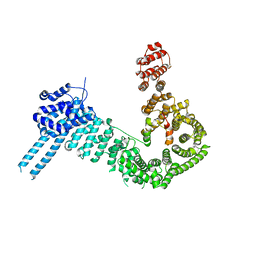 | |
7UPL
 
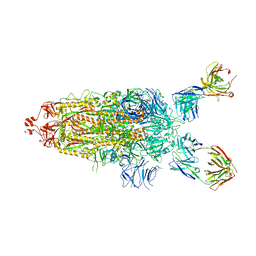 | | SARS-Cov2 Omicron varient S protein structure in complex with neutralizing monoclonal antibody 002-S21F2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, ... | | Authors: | Patel, A, Ortlund, E. | | Deposit date: | 2022-04-15 | | Release date: | 2022-08-10 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Structural insights for neutralization of Omicron variants BA.1, BA.2, BA.4, and BA.5 by a broadly neutralizing SARS-CoV-2 antibody.
Sci Adv, 8, 2022
|
|
7U0P
 
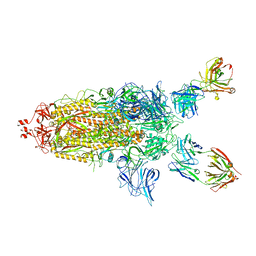 | | SARS-Cov2 S protein structure in complex with neutralizing monoclonal antibody 002-S21F2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, ... | | Authors: | Patel, A, Ortlund, E. | | Deposit date: | 2022-02-18 | | Release date: | 2022-08-10 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.76 Å) | | Cite: | Structural insights for neutralization of Omicron variants BA.1, BA.2, BA.4, and BA.5 by a broadly neutralizing SARS-CoV-2 antibody.
Sci Adv, 8, 2022
|
|
5VQD
 
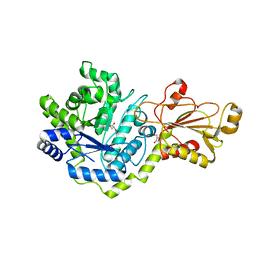 | | Beta-glucoside phosphorylase BglX | | Descriptor: | Beta-glucoside phosphorylase BglX, GLYCEROL | | Authors: | Patel, A, Mark, B.L. | | Deposit date: | 2017-05-08 | | Release date: | 2018-01-17 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural and mechanistic analysis of a beta-glycoside phosphorylase identified by screening a metagenomic library.
J. Biol. Chem., 293, 2018
|
|
5VQE
 
 | | Beta-glucoside phosphorylase BglX bound to 2FGlc | | Descriptor: | 2-deoxy-2-fluoro-alpha-D-glucopyranose, Beta-glucoside phosphorylase BglX | | Authors: | Patel, A, Mark, B.L. | | Deposit date: | 2017-05-08 | | Release date: | 2018-01-17 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.889 Å) | | Cite: | Structural and mechanistic analysis of a beta-glycoside phosphorylase identified by screening a metagenomic library.
J. Biol. Chem., 293, 2018
|
|
2UUL
 
 | | Crystal structure of C-phycocyanin from Phormidium, Lyngbya spp. (Marine) and Spirulina sp. (Fresh water) shows two different ways of energy transfer between two hexamers. | | Descriptor: | BILIVERDINE IX ALPHA, C-PHYCOCYANIN ALPHA CHAIN, C-PHYCOCYANIN BETA CHAIN, ... | | Authors: | Satyanarayana, L, Patel, A, Mishra, S, K Ghosh, P, Suresh, C.G. | | Deposit date: | 2007-03-04 | | Release date: | 2008-05-27 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Crystal Structure of C-Phycocyanin from Phormidium, Lyngbya Spp. (Marine) and Spirulina Sp. (Fresh Water) Shows Two Different Ways of Energy Transfer between Twohexamers.
To be Published
|
|
3J9D
 
 | | Atomic structure of a non-enveloped virus reveals pH sensors for a coordinated process of cell entry | | Descriptor: | Outer capsid protein VP2, ZINC ION | | Authors: | Zhang, X, Patel, A, Celma, C, Roy, P, Zhou, Z.H. | | Deposit date: | 2015-01-09 | | Release date: | 2015-12-09 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Atomic model of a nonenveloped virus reveals pH sensors for a coordinated process of cell entry.
Nat.Struct.Mol.Biol., 23, 2016
|
|
3J9E
 
 | | Atomic structure of a non-enveloped virus reveals pH sensors for a coordinated process of cell entry | | Descriptor: | VP5 | | Authors: | Zhang, X, Patel, A, Celma, C, Roy, P, Zhou, Z.H. | | Deposit date: | 2015-01-10 | | Release date: | 2015-12-09 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Atomic model of a nonenveloped virus reveals pH sensors for a coordinated process of cell entry.
Nat.Struct.Mol.Biol., 23, 2016
|
|
