2YMN
 
 | | Organization of the Influenza Virus Replication Machinery | | Descriptor: | NUCLEOPROTEIN | | Authors: | Moeller, A, Kirchdoerfer, R.N, Potter, C.S, Carragher, B, Wilson, I.A. | | Deposit date: | 2012-10-09 | | Release date: | 2012-12-05 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (20 Å) | | Cite: | Organization of the Influenza Virus Replication Machinery.
Science, 338, 2012
|
|
4ZWJ
 
 | | Crystal structure of rhodopsin bound to arrestin by femtosecond X-ray laser | | Descriptor: | Chimera protein of human Rhodopsin, mouse S-arrestin, and T4 Endolysin | | Authors: | Kang, Y, Zhou, X.E, Gao, X, He, Y, Liu, W, Ishchenko, A, Barty, A, White, T.A, Yefanov, O, Han, G.W, Xu, Q, de Waal, P.W, Ke, J, Tan, M.H.E, Zhang, C, Moeller, A, West, G.M, Pascal, B, Eps, N.V, Caro, L.N, Vishnivetskiy, S.A, Lee, R.J, Suino-Powell, K.M, Gu, X, Pal, K, Ma, J, Zhi, X, Boutet, S, Williams, G.J, Messerschmidt, M, Gati, C, Zatsepin, N.A, Wang, D, James, D, Basu, S, Roy-Chowdhury, S, Conrad, C, Coe, J, Liu, H, Lisova, S, Kupitz, C, Grotjohann, I, Fromme, R, Jiang, Y, Tan, M, Yang, H, Li, J, Wang, M, Zheng, Z, Li, D, Howe, N, Zhao, Y, Standfuss, J, Diederichs, K, Dong, Y, Potter, C.S, Carragher, B, Caffrey, M, Jiang, H, Chapman, H.N, Spence, J.C.H, Fromme, P, Weierstall, U, Ernst, O.P, Katritch, V, Gurevich, V.V, Griffin, P.R, Hubbell, W.L, Stevens, R.C, Cherezov, V, Melcher, K, Xu, H.E, GPCR Network (GPCR) | | Deposit date: | 2015-05-19 | | Release date: | 2015-07-29 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (3.302 Å) | | Cite: | Crystal structure of rhodopsin bound to arrestin by femtosecond X-ray laser.
Nature, 523, 2015
|
|
7ZU0
 
 | | HOPS tethering complex from yeast | | Descriptor: | E3 ubiquitin-protein ligase PEP5, Vacuolar membrane protein PEP3, Vacuolar morphogenesis protein 6, ... | | Authors: | Shvarev, D, Schoppe, J, Koenig, C, Perz, A, Fuellbrunn, N, Kiontke, S, Langemeyer, L, Januliene, D, Schnelle, K, Kuemmel, D, Froehlich, F, Moeller, A, Ungermann, C. | | Deposit date: | 2022-05-11 | | Release date: | 2022-09-28 | | Last modified: | 2025-04-16 | | Method: | ELECTRON MICROSCOPY (4.4 Å) | | Cite: | Structure of the HOPS tethering complex, a lysosomal membrane fusion machinery.
Elife, 11, 2022
|
|
8OSH
 
 | |
8OSF
 
 | | AAA+ motor subunit ChlI of magnesium chelatase, hexamer conformation A | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Shvarev, D, Moeller, A. | | Deposit date: | 2023-04-18 | | Release date: | 2023-09-06 | | Last modified: | 2023-11-29 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Conformational variability of cyanobacterial ChlI, the AAA+ motor of magnesium chelatase involved in chlorophyll biosynthesis.
Mbio, 14, 2023
|
|
8OSG
 
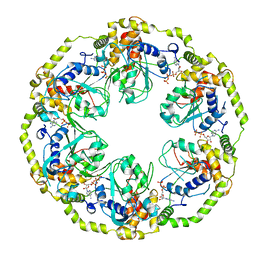 | | AAA+ motor subunit ChlI of magnesium chelatase, hexamer conformation B | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Shvarev, D, Moeller, A. | | Deposit date: | 2023-04-18 | | Release date: | 2023-09-06 | | Last modified: | 2023-11-29 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Conformational variability of cyanobacterial ChlI, the AAA+ motor of magnesium chelatase involved in chlorophyll biosynthesis.
Mbio, 14, 2023
|
|
6ENY
 
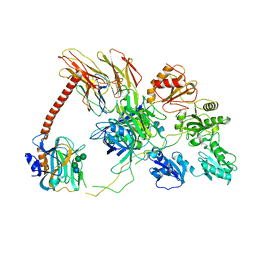 | | Structure of the human PLC editing module | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Beta-2-microglobulin, Calreticulin, ... | | Authors: | Trowitzsch, S, Januliene, D, Blees, A, Moeller, A, Tampe, R. | | Deposit date: | 2017-10-07 | | Release date: | 2017-11-29 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (5.8 Å) | | Cite: | Structure of the human MHC-I peptide-loading complex.
Nature, 551, 2017
|
|
8C80
 
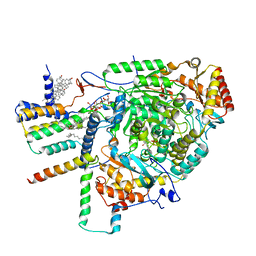 | | Cryo-EM structure of the yeast SPT-Orm1-Monomer complex | | Descriptor: | 2-{[(4-O-alpha-D-glucopyranosyl-alpha-D-glucopyranosyl)oxy]methyl}-4-{[(3beta,9beta,14beta,17beta,25R)-spirost-5-en-3-yl]oxy}butyl 4-O-alpha-D-glucopyranosyl-alpha-D-glucopyranoside, ERGOSTEROL, N-[(2S,3S,4R)-1,3,4-trihydroxyoctadecan-2-yl]hexacosanamide, ... | | Authors: | Schaefer, J, Koerner, C, Parey, K, Januliene, D, Moeller, A, Froehlich, F. | | Deposit date: | 2023-01-18 | | Release date: | 2023-10-11 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structure of the ceramide-bound SPOTS complex.
Nat Commun, 14, 2023
|
|
8C81
 
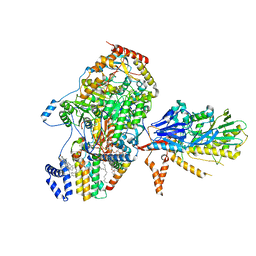 | | Cryo-EM structure of the yeast SPT-Orm1-Sac1 complex | | Descriptor: | 2-{[(4-O-alpha-D-glucopyranosyl-alpha-D-glucopyranosyl)oxy]methyl}-4-{[(3beta,9beta,14beta,17beta,25R)-spirost-5-en-3-yl]oxy}butyl 4-O-alpha-D-glucopyranosyl-alpha-D-glucopyranoside, ERGOSTEROL, N-[(2S,3S,4R)-1,3,4-trihydroxyoctadecan-2-yl]hexacosanamide, ... | | Authors: | Schaefer, J, Koerner, C, Parey, K, Januliene, D, Moeller, A, Froehlich, F. | | Deposit date: | 2023-01-18 | | Release date: | 2023-10-11 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structure of the ceramide-bound SPOTS complex.
Nat Commun, 14, 2023
|
|
8C82
 
 | | Cryo-EM structure of the yeast SPT-Orm1-Dimer complex | | Descriptor: | 2-{[(4-O-alpha-D-glucopyranosyl-alpha-D-glucopyranosyl)oxy]methyl}-4-{[(3beta,9beta,14beta,17beta,25R)-spirost-5-en-3-yl]oxy}butyl 4-O-alpha-D-glucopyranosyl-alpha-D-glucopyranoside, N-[(2S,3S,4R)-1,3,4-trihydroxyoctadecan-2-yl]hexacosanamide, PYRIDOXAL-5'-PHOSPHATE, ... | | Authors: | Schaefer, J, Koerner, C, Parey, K, Januliene, D, Moeller, A, Froehlich, F. | | Deposit date: | 2023-01-18 | | Release date: | 2023-10-11 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structure of the ceramide-bound SPOTS complex.
Nat Commun, 14, 2023
|
|
7PH3
 
 | | AMP-PNP bound nanodisc reconstituted MsbA with nanobodies, spin-labeled at position A60C | | Descriptor: | (1~{R},4~{R},11~{S},14~{S},19~{Z})-19-[2-[2,5-bis(oxidanylidene)pyrrolidin-1-yl]ethylimino]-7,8,17,18-tetraoxa-1,4,11,14-tetrazatricyclo[12.6.2.2^{4,11}]tetracosane-6,9,16-trione, 1,2-Distearoyl-sn-glycerophosphoethanolamine, ATP-dependent lipid A-core flippase, ... | | Authors: | Parey, K, Januliene, D, Galazzo, L, Meier, G, Vecchis, D, Striednig, B, Hilbi, H, Schaefer, L.V, Kuprov, I, Bordignon, E, Seeger, M.A, Moeller, A. | | Deposit date: | 2021-08-16 | | Release date: | 2022-08-24 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | The ABC transporter MsbA adopts the wide inward-open conformation in E. coli cells.
Sci Adv, 8, 2022
|
|
7PH2
 
 | | Nanodisc reconstituted MsbA in complex with nanobodies, spin-labeled at position A60C | | Descriptor: | (1~{R},4~{R},11~{S},14~{S},19~{Z})-19-[2-[2,5-bis(oxidanylidene)pyrrolidin-1-yl]ethylimino]-7,8,17,18-tetraoxa-1,4,11,14-tetrazatricyclo[12.6.2.2^{4,11}]tetracosane-6,9,16-trione, (2~{R},4~{R},5~{R},6~{R})-6-[(1~{R})-1,2-bis(oxidanyl)ethyl]-4-[(2~{R},3~{S},4~{S},5~{R},6~{R})-6-[(1~{S})-1,2-bis(oxidanyl)ethyl]-4-[(2~{R},3~{S},4~{S},5~{S},6~{R})-6-[(1~{S})-1,2-bis(oxidanyl)ethyl]-3,4,5-tris(oxidanyl)oxan-2-yl]oxy-3,5-bis(oxidanyl)oxan-2-yl]oxy-2-[[(2~{R},3~{S},4~{R},5~{R},6~{R})-4-[(3~{R})-3-nonanoyloxytetradecanoyl]oxy-5-[[(3~{R})-3-octanoyloxytetradecanoyl]amino]-6-[[(2~{R},3~{S},4~{S},5~{S},6~{R})-3-oxidanyl-5-[[(3~{R})-3-oxidanylnonanoyl]amino]-4-[(3~{R})-3-oxidanyltetradecanoyl]oxy-6-phosphonooxy-oxan-2-yl]methoxy]-3-phosphonooxy-oxan-2-yl]methoxy]-5-oxidanyl-oxane-2-carboxylic acid, ATP-dependent lipid A-core flippase, ... | | Authors: | Januliene, D, Parey, K, Galazzo, L, Meier, G, Vecchis, D, Striednig, B, Hilbi, H, Schaefer, L.V, Kuprov, I, Bordignon, E, Seeger, M.A, Moeller, A. | | Deposit date: | 2021-08-16 | | Release date: | 2022-08-24 | | Last modified: | 2025-07-09 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | The ABC transporter MsbA adopts the wide inward-open conformation in E. coli cells.
Sci Adv, 8, 2022
|
|
7PH7
 
 | | Nanodisc reconstituted MsbA in complex with nanobodies, spin-labeled at position T68C | | Descriptor: | (1~{R},4~{R},11~{S},14~{S},19~{Z})-19-[2-[2,5-bis(oxidanylidene)pyrrolidin-1-yl]ethylimino]-7,8,17,18-tetraoxa-1,4,11,14-tetrazatricyclo[12.6.2.2^{4,11}]tetracosane-6,9,16-trione, (2~{R},4~{R},5~{R},6~{R})-6-[(1~{R})-1,2-bis(oxidanyl)ethyl]-4-[(2~{R},3~{S},4~{S},5~{R},6~{R})-6-[(1~{S})-1,2-bis(oxidanyl)ethyl]-4-[(2~{R},3~{S},4~{S},5~{S},6~{R})-6-[(1~{S})-1,2-bis(oxidanyl)ethyl]-3,4,5-tris(oxidanyl)oxan-2-yl]oxy-3,5-bis(oxidanyl)oxan-2-yl]oxy-2-[[(2~{R},3~{S},4~{R},5~{R},6~{R})-4-[(3~{R})-3-nonanoyloxytetradecanoyl]oxy-5-[[(3~{R})-3-octanoyloxytetradecanoyl]amino]-6-[[(2~{R},3~{S},4~{S},5~{S},6~{R})-3-oxidanyl-5-[[(3~{R})-3-oxidanylnonanoyl]amino]-4-[(3~{R})-3-oxidanyltetradecanoyl]oxy-6-phosphonooxy-oxan-2-yl]methoxy]-3-phosphonooxy-oxan-2-yl]methoxy]-5-oxidanyl-oxane-2-carboxylic acid, ATP-binding transport protein multicopy suppressor of htrB, ... | | Authors: | Parey, K, Januliene, D, Galazzo, L, Meier, G, Vecchis, D, Striednig, B, Hilbi, H, Schaefer, L.V, Kuprov, I, Bordignon, E, Seeger, M.A, Moeller, A. | | Deposit date: | 2021-08-16 | | Release date: | 2022-08-24 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | The ABC transporter MsbA adopts the wide inward-open conformation in E. coli cells.
Sci Adv, 8, 2022
|
|
8QX8
 
 | | Endosomal membrane tethering complex CORVET | | Descriptor: | E3 ubiquitin-protein ligase PEP5, Vacuolar membrane protein PEP3, Vacuolar protein sorting-associated protein 16, ... | | Authors: | Shvarev, D, Ungermann, C, Moeller, A. | | Deposit date: | 2023-10-23 | | Release date: | 2024-07-03 | | Last modified: | 2025-07-09 | | Method: | ELECTRON MICROSCOPY (4.6 Å) | | Cite: | Structure of the endosomal CORVET tethering complex.
Nat Commun, 15, 2024
|
|
8AVY
 
 | | The ABCB1 L335C mutant (mABCB1) in the Apo state | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, ATP-dependent translocase ABCB1, CHOLESTEROL HEMISUCCINATE, ... | | Authors: | Parey, K, Januliene, D, Gewering, T, Moeller, A. | | Deposit date: | 2022-08-27 | | Release date: | 2023-09-06 | | Last modified: | 2025-07-09 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Tracing the substrate translocation mechanism in P-glycoprotein.
Elife, 12, 2024
|
|
8PEE
 
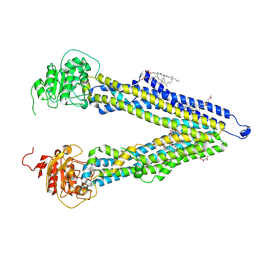 | | ABCB1 L335C mutant (mABCB1) in the inward facing state bound to AAC | | Descriptor: | (4S,11S,18S)-4-[[(2,4-dinitrophenyl)disulfanyl]methyl]-11,18-dimethyl-6,13,20-trithia-3,10,17,22,23,24-hexazatetracyclo[17.2.1.1^{5,8}.1^{12,15}]tetracosa-1(21),5(24),7,12(23),14,19(22)-hexaene-2,9,16-trione, (4~{S},11~{S},18~{S})-4,11-dimethyl-18-(sulfanylmethyl)-6,13,20-trithia-3,10,17,22,23,24-hexazatetracyclo[17.2.1.1^{5,8}.1^{12,15}]tetracosa-1(21),5(24),7,12(23),14,19(22)-hexaene-2,9,16-trione, ATP-dependent translocase ABCB1, ... | | Authors: | Parey, K, Januliene, D, Gewering, T, Moeller, A. | | Deposit date: | 2023-06-13 | | Release date: | 2024-03-20 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Tracing the substrate translocation mechanism in P-glycoprotein.
Elife, 12, 2024
|
|
8QOG
 
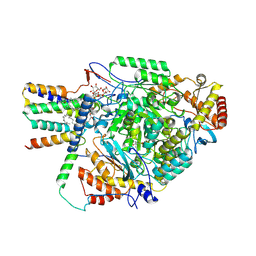 | | Cryo-EM structure of the yeast SPT-Orm2-Monomer complex | | Descriptor: | 2-{[(4-O-alpha-D-glucopyranosyl-alpha-D-glucopyranosyl)oxy]methyl}-4-{[(3beta,9beta,14beta,17beta,25R)-spirost-5-en-3-yl]oxy}butyl 4-O-alpha-D-glucopyranosyl-alpha-D-glucopyranoside, ORM2 isoform 1, PYRIDOXAL-5'-PHOSPHATE, ... | | Authors: | Schaefer, J, Koerner, C, Moeller, A, Froehlich, F. | | Deposit date: | 2023-09-28 | | Release date: | 2024-09-04 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | The structure of the Orm2-containing serine palmitoyltransferase complex reveals distinct inhibitory potentials of yeast Orm proteins.
Cell Rep, 43, 2024
|
|
8QOF
 
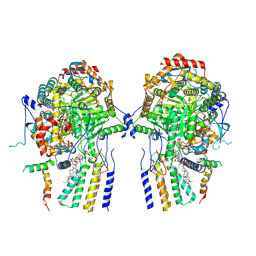 | | Cryo-EM structure of the yeast SPT-Orm2-Dimer complex | | Descriptor: | 2-{[(4-O-alpha-D-glucopyranosyl-alpha-D-glucopyranosyl)oxy]methyl}-4-{[(3beta,9beta,14beta,17beta,25R)-spirost-5-en-3-yl]oxy}butyl 4-O-alpha-D-glucopyranosyl-alpha-D-glucopyranoside, ORM2 isoform 1, PYRIDOXAL-5'-PHOSPHATE, ... | | Authors: | Schaefer, J, Koerner, C, Moeller, A, Froehlich, F. | | Deposit date: | 2023-09-28 | | Release date: | 2024-09-04 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | The structure of the Orm2-containing serine palmitoyltransferase complex reveals distinct inhibitory potentials of yeast Orm proteins.
Cell Rep, 43, 2024
|
|
8QTN
 
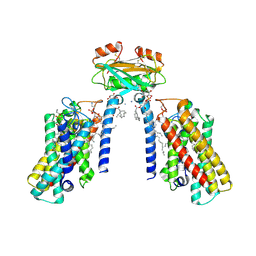 | | Cryo-EM structure of the apo yeast Ceramide Synthase | | Descriptor: | 1,2-Distearoyl-sn-glycerophosphoethanolamine, AMMONIUM ION, Ceramide synthase LAC1, ... | | Authors: | Schaefer, J, Clausmeyer, L, Koerner, C, Moeller, A, Froehlich, F. | | Deposit date: | 2023-10-12 | | Release date: | 2024-10-23 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structure of the yeast ceramide synthase.
Nat.Struct.Mol.Biol., 32, 2025
|
|
8QTR
 
 | | Cryo-EM structure of the FB-bound yeast Ceramide Synthase | | Descriptor: | 1,2-Distearoyl-sn-glycerophosphoethanolamine, AMMONIUM ION, Ceramide synthase LAC1, ... | | Authors: | Schaefer, J, Clausmeyer, L, Koerner, C, Moeller, A, Froehlich, F. | | Deposit date: | 2023-10-13 | | Release date: | 2024-10-23 | | Last modified: | 2025-07-09 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structure of the yeast ceramide synthase.
Nat.Struct.Mol.Biol., 32, 2025
|
|
8C7G
 
 | | Drosophila melanogaster Rab7 GEF complex Mon1-Ccz1-Bulli | | Descriptor: | Caffeine, calcium, zinc sensitivity 1, ... | | Authors: | Schaefer, J, Herrmann, E, Kuemmel, D, Moeller, A. | | Deposit date: | 2023-01-15 | | Release date: | 2023-05-17 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structure of the metazoan Rab7 GEF complex Mon1-Ccz1-Bulli.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
7PH4
 
 | | AMP-PNP bound nanodisc reconstituted MsbA with nanobodies, spin-labeled at position T68C | | Descriptor: | (1~{R},4~{R},11~{S},14~{S},19~{Z})-19-[2-[2,5-bis(oxidanylidene)pyrrolidin-1-yl]ethylimino]-7,8,17,18-tetraoxa-1,4,11,14-tetrazatricyclo[12.6.2.2^{4,11}]tetracosane-6,9,16-trione, ATP-dependent lipid A-core flippase, DODECYL-BETA-D-MALTOSIDE, ... | | Authors: | Parey, K, Januliene, D, Galazzo, L, Meier, G, Vecchis, D, Striednig, B, Hilbi, H, Schaefer, L.V, Kuprov, I, Bordignon, E, Seeger, M.A, Moeller, A. | | Deposit date: | 2021-08-16 | | Release date: | 2022-08-24 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | The ABC transporter MsbA adopts the wide inward-open conformation in E. coli cells.
Sci Adv, 8, 2022
|
|
7ZK6
 
 | | ABCB1 L335C mutant (mABCB1) in the outward facing state bound to 2 molecules of AAC | | Descriptor: | (4S,11S,18S)-4-[[(2,4-dinitrophenyl)disulfanyl]methyl]-11,18-dimethyl-6,13,20-trithia-3,10,17,22,23,24-hexazatetracyclo[17.2.1.1^{5,8}.1^{12,15}]tetracosa-1(21),5(24),7,12(23),14,19(22)-hexaene-2,9,16-trione, (4~{S},11~{S},18~{S})-4,11-dimethyl-18-(sulfanylmethyl)-6,13,20-trithia-3,10,17,22,23,24-hexazatetracyclo[17.2.1.1^{5,8}.1^{12,15}]tetracosa-1(21),5(24),7,12(23),14,19(22)-hexaene-2,9,16-trione, ADENOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Parey, K, Januliene, D, Gewering, T, Moeller, A. | | Deposit date: | 2022-04-12 | | Release date: | 2023-04-26 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Tracing the substrate translocation mechanism in P-glycoprotein.
Elife, 12, 2024
|
|
7ZKA
 
 | | ABCB1 V978C mutant (mABCB1) in the outward facing state bound to AAC | | Descriptor: | (4~{S},11~{S},18~{S})-4,11-dimethyl-18-(sulfanylmethyl)-6,13,20-trithia-3,10,17,22,23,24-hexazatetracyclo[17.2.1.1^{5,8}.1^{12,15}]tetracosa-1(21),5(24),7,12(23),14,19(22)-hexaene-2,9,16-trione, ADENOSINE-5'-TRIPHOSPHATE, ATP-dependent translocase ABCB1, ... | | Authors: | Parey, K, Januliene, D, Gewering, T, Urbatsch, I, Zhang, Q, Moeller, A. | | Deposit date: | 2022-04-12 | | Release date: | 2023-04-26 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Tracing the substrate translocation mechanism in P-glycoprotein.
Elife, 12, 2024
|
|
7ZK9
 
 | | ABCB1 L971C mutant (mABCB1) in the inward facing state | | Descriptor: | (4~{S},11~{S},18~{S})-4,11-dimethyl-18-(sulfanylmethyl)-6,13,20-trithia-3,10,17,22,23,24-hexazatetracyclo[17.2.1.1^{5,8}.1^{12,15}]tetracosa-1(21),5(24),7,12(23),14,19(22)-hexaene-2,9,16-trione, ATP-dependent translocase ABCB1 | | Authors: | Parey, K, Januliene, D, Gewering, T, Zhang, Q, Moeller, A. | | Deposit date: | 2022-04-12 | | Release date: | 2023-04-26 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Tracing the substrate translocation mechanism in P-glycoprotein.
Elife, 12, 2024
|
|
