1UZG
 
 | | CRYSTAL STRUCTURE OF THE DENGUE TYPE 3 VIRUS ENVELOPE PROTEIN | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, MAJOR ENVELOPE PROTEIN E, alpha-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[beta-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Modis, Y, Harrison, S.C. | | Deposit date: | 2004-03-11 | | Release date: | 2005-03-15 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Variable Surface Epitopes in the Crystal Structure of Dengue Virus Type 3 Envelope Glycoprotein
J.Virol., 79, 2005
|
|
1OK8
 
 | |
1DLU
 
 | | UNLIGANDED BIOSYNTHETIC THIOLASE FROM ZOOGLOEA RAMIGERA | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, BIOSYNTHETIC THIOLASE, SULFATE ION | | Authors: | Modis, Y, Wierenga, R.K. | | Deposit date: | 1999-12-12 | | Release date: | 2000-04-24 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Crystallographic analysis of the reaction pathway of Zoogloea ramigera biosynthetic thiolase.
J.Mol.Biol., 297, 2000
|
|
1DM3
 
 | |
1DLV
 
 | |
1QFL
 
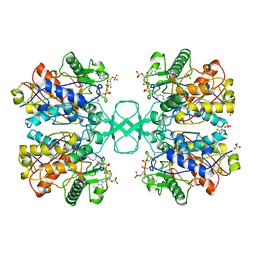 | |
1OAN
 
 | |
1OKE
 
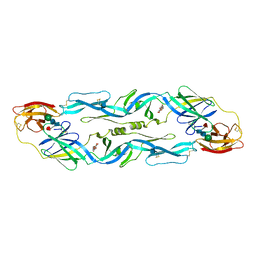 | |
1DCI
 
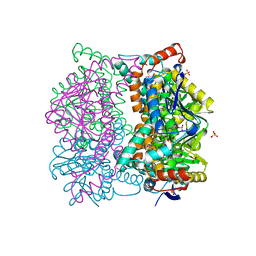 | | DIENOYL-COA ISOMERASE | | Descriptor: | 1,2-ETHANEDIOL, DIENOYL-COA ISOMERASE, MAGNESIUM ION, ... | | Authors: | Modis, Y, Filppula, S.A, Novikov, D, Norledge, B, Hiltunen, J.K, Wierenga, R.K. | | Deposit date: | 1998-02-13 | | Release date: | 1999-03-30 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | The crystal structure of dienoyl-CoA isomerase at 1.5 A resolution reveals the importance of aspartate and glutamate sidechains for catalysis.
Structure, 6, 1998
|
|
1I31
 
 | | MU2 ADAPTIN SUBUNIT (AP50) OF AP2 CLATHRIN ADAPTOR, COMPLEXED WITH EGFR INTERNALIZATION PEPTIDE FYRALM AT 2.5 A RESOLUTION | | Descriptor: | CLATHRIN COAT ASSEMBLY PROTEIN AP50, EPIDERMAL GROWTH FACTOR RECEPTOR | | Authors: | Modis, Y, Boll, W, Rapoport, I, Kirchhausen, T. | | Deposit date: | 2001-02-12 | | Release date: | 2001-02-21 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | MU2 ADAPTIN SUBUNIT (AP50) OF AP2 CLATHRIN ADAPTOR, COMPLEXED WITH EGFR INTERNALIZATION PEPTIDE FYRALM AT 2.5 A RESOLUTION
To be Published
|
|
3J0A
 
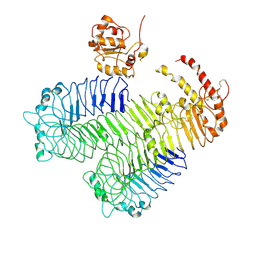 | | Homology model of human Toll-like receptor 5 fitted into an electron microscopy single particle reconstruction | | Descriptor: | Toll-like receptor 5, alpha-L-fucopyranose-(1-6)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Modis, Y, Zhou, K, Kanai, R, Lee, P, Wang, H.W. | | Deposit date: | 2011-06-02 | | Release date: | 2011-12-28 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (26 Å) | | Cite: | Toll-like receptor 5 forms asymmetric dimers in the absence of flagellin.
J.Struct.Biol., 177, 2012
|
|
3G7T
 
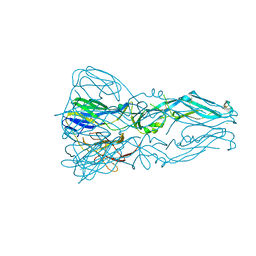 | |
4ILD
 
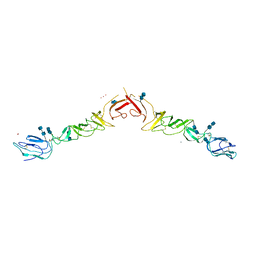 | | Crystal structure of truncated Bovine viral diarrhea virus 1 E2 envelope protein | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, Envelope glycoprotein E2, ... | | Authors: | Modis, Y, Li, Y, Wang, J. | | Deposit date: | 2012-12-30 | | Release date: | 2013-04-10 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.27 Å) | | Cite: | Crystal structure of glycoprotein E2 from bovine viral diarrhea virus.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
7A4A
 
 | |
6TL1
 
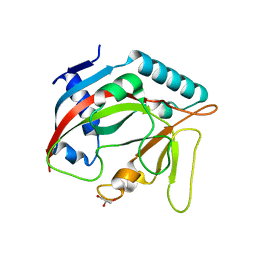 | | Crystal structure of the TASOR pseudo-PARP domain | | Descriptor: | GLYCEROL, Protein TASOR | | Authors: | Douse, C.H, Timms, R.T, Freund, S.M.V, Modis, Y. | | Deposit date: | 2019-11-29 | | Release date: | 2020-09-16 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | TASOR is a pseudo-PARP that directs HUSH complex assembly and epigenetic transposon control.
Nat Commun, 11, 2020
|
|
2I69
 
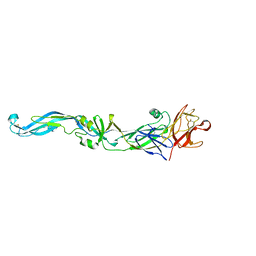 | | Crystal structure of the West Nile virus envelope glycoprotein | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[beta-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, Polyprotein | | Authors: | Kanai, R, Modis, Y. | | Deposit date: | 2006-08-28 | | Release date: | 2006-11-21 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.11 Å) | | Cite: | Crystal structure of west nile virus envelope glycoprotein reveals viral surface epitopes.
J.Virol., 80, 2006
|
|
5OFA
 
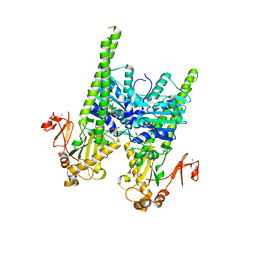 | | Crystal structure of human MORC2 (residues 1-603) with spinal muscular atrophy mutation T424R | | Descriptor: | MAGNESIUM ION, MORC family CW-type zinc finger protein 2, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, ... | | Authors: | Douse, C.H, Liu, Y, Modis, Y. | | Deposit date: | 2017-07-10 | | Release date: | 2018-02-14 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.57 Å) | | Cite: | Neuropathic MORC2 mutations perturb GHKL ATPase dimerization dynamics and epigenetic silencing by multiple structural mechanisms.
Nat Commun, 9, 2018
|
|
5OFB
 
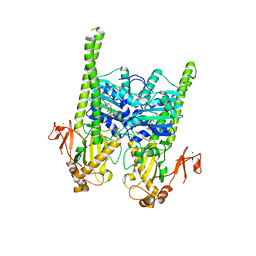 | | Crystal structure of human MORC2 (residues 1-603) with spinal muscular atrophy mutation S87L | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, MORC family CW-type zinc finger protein 2, ... | | Authors: | Douse, C.H, Liu, Y, Modis, Y. | | Deposit date: | 2017-07-10 | | Release date: | 2018-02-14 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Neuropathic MORC2 mutations perturb GHKL ATPase dimerization dynamics and epigenetic silencing by multiple structural mechanisms.
Nat Commun, 9, 2018
|
|
5OF9
 
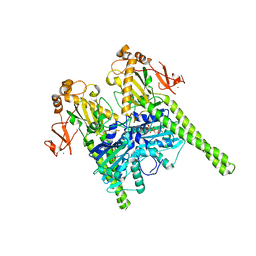 | | Crystal structure of human MORC2 (residues 1-603) | | Descriptor: | MAGNESIUM ION, MORC family CW-type zinc finger protein 2, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, ... | | Authors: | Douse, C.H, Shamin, M, Modis, Y. | | Deposit date: | 2017-07-10 | | Release date: | 2018-02-14 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.807 Å) | | Cite: | Neuropathic MORC2 mutations perturb GHKL ATPase dimerization dynamics and epigenetic silencing by multiple structural mechanisms.
Nat Commun, 9, 2018
|
|
7Z36
 
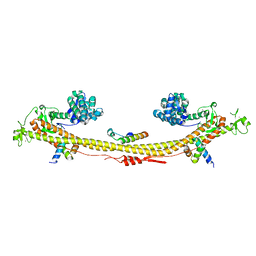 | | Crystal structure of the KAP1 tripartite motif in complex with the ZNF93 KRAB domain | | Descriptor: | Endolysin,Transcription intermediary factor 1-beta,Isoform 2 of Transcription intermediary factor 1-beta, SMARCAD1 CUE1 domain, ZINC ION, ... | | Authors: | Stoll, G.A, Modis, Y. | | Deposit date: | 2022-03-01 | | Release date: | 2022-11-02 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure and functional mapping of the KRAB-KAP1 repressor complex.
Embo J., 41, 2022
|
|
6SWG
 
 | |
5ELX
 
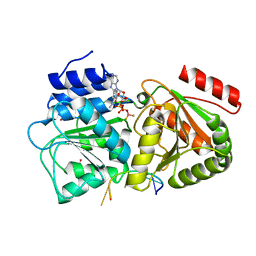 | | S. cerevisiae Dbp5 bound to RNA and mant-ADP BeF3 | | Descriptor: | ATP-dependent RNA helicase DBP5, BERYLLIUM TRIFLUORIDE ION, MAGNESIUM ION, ... | | Authors: | Merchant, M.K, Modis, Y. | | Deposit date: | 2015-11-05 | | Release date: | 2016-02-24 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Pi Release Limits the Intrinsic and RNA-Stimulated ATPase Cycles of DEAD-Box Protein 5 (Dbp5).
J.Mol.Biol., 428, 2016
|
|
3NSW
 
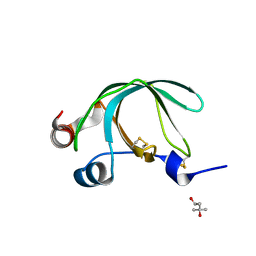 | | Crystal Structure of Ancylostoma ceylanicum Excretory-Secretory Protein 2 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Excretory-secretory protein 2 | | Authors: | Kucera, K, Modis, Y. | | Deposit date: | 2010-07-02 | | Release date: | 2011-05-18 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Ancylostoma ceylanicum Excretory-Secretory Protein 2 Adopts a Netrin-Like Fold and Defines a Novel Family of Nematode Proteins.
J.Mol.Biol., 408, 2011
|
|
6QAJ
 
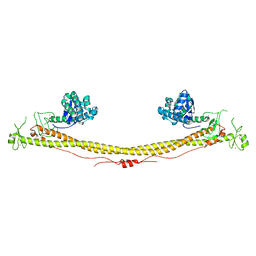 | | Structure of the tripartite motif of KAP1/TRIM28 | | Descriptor: | Endolysin,Transcription intermediary factor 1-beta, ZINC ION | | Authors: | Stoll, G.A, Oda, S, Yu, M, Modis, Y. | | Deposit date: | 2018-12-19 | | Release date: | 2019-07-24 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.901 Å) | | Cite: | Structure of KAP1 tripartite motif identifies molecular interfaces required for retroelement silencing.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
3TS9
 
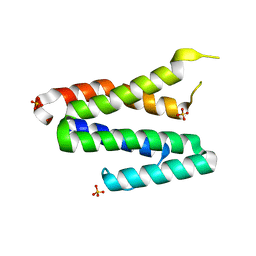 | | Crystal Structure of the MDA5 Helicase Insert Domain | | Descriptor: | Interferon-induced helicase C domain-containing protein 1, SULFATE ION | | Authors: | Berke, I.C, Modis, Y. | | Deposit date: | 2011-09-12 | | Release date: | 2012-02-22 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.003 Å) | | Cite: | MDA5 cooperatively forms dimers and ATP-sensitive filaments upon binding double-stranded RNA.
Embo J., 31, 2012
|
|
