4E3R
 
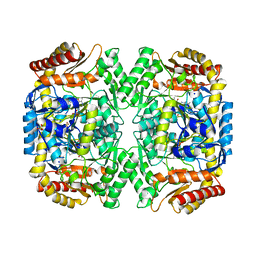 | | PLP-bound aminotransferase mutant crystal structure from Vibrio fluvialis | | Descriptor: | Pyruvate transaminase, SODIUM ION, SULFATE ION | | Authors: | Midelfort, K.S, Kumar, R, Han, S, Karmilowicz, M.J, McConnell, K, Gehlhaar, D.K, Mistry, A, Chang, J.S, Anderson, M, Vilalobos, A, Minshull, J, Govindarajan, S, Wong, J.W. | | Deposit date: | 2012-03-10 | | Release date: | 2012-10-10 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Redesigning and characterizing the substrate specificity and activity of Vibrio fluvialis aminotransferase for the synthesis of imagabalin.
Protein Eng.Des.Sel., 26, 2013
|
|
4E3Q
 
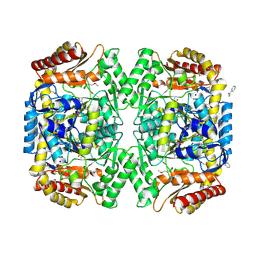 | | PMP-bound form of Aminotransferase crystal structure from Vibrio fluvialis | | Descriptor: | 4'-DEOXY-4'-AMINOPYRIDOXAL-5'-PHOSPHATE, BENZAMIDINE, Pyruvate transaminase, ... | | Authors: | Midelfort, K.S, Kumar, R, Han, S, Karmilowicz, M.J, McConnell, K, Gehlhaar, D.K, Mistry, A, Chang, J.S, Anderson, M, Vilalobos, A, Minshull, J, Govindarajan, S, Wong, J.W. | | Deposit date: | 2012-03-10 | | Release date: | 2012-10-10 | | Last modified: | 2013-01-02 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Redesigning and characterizing the substrate specificity and activity of Vibrio fluvialis aminotransferase for the synthesis of imagabalin.
Protein Eng.Des.Sel., 26, 2013
|
|
1D7O
 
 | | CRYSTAL STRUCTURE OF BRASSICA NAPUS ENOYL ACYL CARRIER PROTEIN REDUCTASE COMPLEXED WITH NAD AND TRICLOSAN | | Descriptor: | ENOYL-[ACYL-CARRIER PROTEIN] REDUCTASE (NADH) PRECURSOR, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, TRICLOSAN | | Authors: | Roujeinikova, A, Levy, C, Rowsell, S, Sedelnikova, S, Baker, P.J, Minshull, C.A, Mistry, A, Colls, J.G, Camble, R, Stuitje, A.R, Slabas, A.R, Rafferty, J.B, Pauptit, R.A, Viner, R, Rice, D.W. | | Deposit date: | 1999-10-19 | | Release date: | 1999-11-08 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystallographic analysis of triclosan bound to enoyl reductase.
J.Mol.Biol., 294, 1999
|
|
2ABJ
 
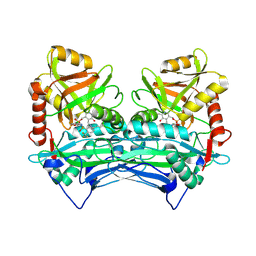 | | Crystal structure of human branched chain amino acid transaminase in a complex with an inhibitor, C16H10N2O4F3SCl, and pyridoxal 5' phosphate. | | Descriptor: | Branched-chain-amino-acid aminotransferase, cytosolic, N'-(5-CHLOROBENZOFURAN-2-CARBONYL)-2-(TRIFLUOROMETHYL)BENZENESULFONOHYDRAZIDE, ... | | Authors: | Ohren, J.F, Moreland, D.W, Rubin, J.R, Hu, H.L, McConnell, P.C, Mistry, A, Mueller, W.T, Scholten, J.D, Hasemann, C.H. | | Deposit date: | 2005-07-15 | | Release date: | 2006-06-27 | | Last modified: | 2017-10-11 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The design and synthesis of human branched-chain amino acid aminotransferase inhibitors for treatment of neurodegenerative diseases.
Bioorg.Med.Chem.Lett., 16, 2006
|
|
3H3C
 
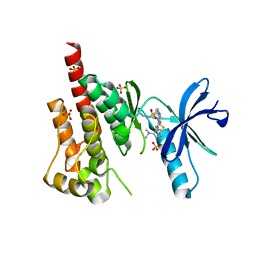 | | Crystal structure of PYK2 in complex with Sulfoximine-substituted trifluoromethylpyrimidine analog | | Descriptor: | 4-{[4-{[(1R,2R)-2-(dimethylamino)cyclopentyl]amino}-5-(trifluoromethyl)pyrimidin-2-yl]amino}-N-methylbenzenesulfonamide, Protein tyrosine kinase 2 beta, SULFATE ION | | Authors: | Han, S, Mistry, A. | | Deposit date: | 2009-04-16 | | Release date: | 2009-05-26 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Sulfoximine-substituted trifluoromethylpyrimidine analogs as inhibitors of proline-rich tyrosine kinase 2 (PYK2) show reduced hERG activity.
Bioorg.Med.Chem.Lett., 19, 2009
|
|
3HZS
 
 | | S. aureus monofunctional glycosyltransferase (MtgA)in complex with moenomycin | | Descriptor: | MOENOMYCIN, Monofunctional glycosyltransferase, PHOSPHATE ION | | Authors: | Heaslet, H, Miller, A.A, Shaw, B, Mistry, A. | | Deposit date: | 2009-06-24 | | Release date: | 2009-07-07 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Characterization of the active site of S. aureus monofunctional glycosyltransferase (Mtg) by site-directed mutation and structural analysis of the protein complexed with moenomycin
J.Struct.Biol., 167, 2009
|
|
3ET7
 
 | | Crystal structure of PYK2 complexed with PF-2318841 | | Descriptor: | 5-{[4-{[2-(pyrrolidin-1-ylsulfonyl)benzyl]amino}-5-(trifluoromethyl)pyrimidin-2-yl]amino}-1,3-dihydro-2H-indol-2-one, PHOSPHATE ION, Protein tyrosine kinase 2 beta | | Authors: | Han, S. | | Deposit date: | 2008-10-07 | | Release date: | 2009-06-23 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Trifluoromethylpyrimidine-based inhibitors of proline-rich tyrosine kinase 2 (PYK2): structure-activity relationships and strategies for the elimination of reactive metabolite formation.
Bioorg.Med.Chem.Lett., 18, 2008
|
|
3O1G
 
 | |
3O0U
 
 | |
1A2N
 
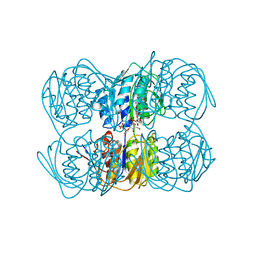 | | STRUCTURE OF THE C115A MUTANT OF MURA COMPLEXED WITH THE FLUORINATED ANALOG OF THE REACTION TETRAHEDRAL INTERMEDIATE | | Descriptor: | UDP-N-ACETYLGLUCOSAMINE ENOLPYRUVYL TRANSFERASE, URIDINE-DIPHOSPHATE-2(N-ACETYLGLUCOSAMINYL-3-FLUORO-2-PHOSPHONOOXY)PROPIONIC ACID | | Authors: | Skarzynski, T. | | Deposit date: | 1998-01-06 | | Release date: | 1998-04-29 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Stereochemical course of enzymatic enolpyruvyl transfer and catalytic conformation of the active site revealed by the crystal structure of the fluorinated analogue of the reaction tetrahedral intermediate bound to the active site of the C115A mutant of MurA
Biochemistry, 37, 1998
|
|
1UAE
 
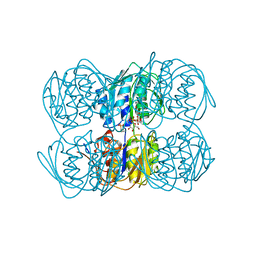 | | STRUCTURE OF UDP-N-ACETYLGLUCOSAMINE ENOLPYRUVYL TRANSFERASE | | Descriptor: | UDP-N-ACETYLGLUCOSAMINE ENOLPYRUVYL TRANSFERASE, URIDINE-DIPHOSPHATE-N-ACETYLGLUCOSAMINE, [(1R)-1-hydroxypropyl]phosphonic acid | | Authors: | Skarzynski, T. | | Deposit date: | 1996-09-30 | | Release date: | 1997-09-04 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure of UDP-N-acetylglucosamine enolpyruvyl transferase, an enzyme essential for the synthesis of bacterial peptidoglycan, complexed with substrate UDP-N-acetylglucosamine and the drug fosfomycin.
Structure, 4, 1996
|
|
2OBD
 
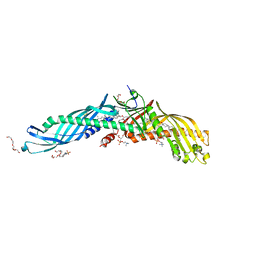 | | Crystal Structure of Cholesteryl Ester Transfer Protein | | Descriptor: | 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ... | | Authors: | Qiu, X. | | Deposit date: | 2006-12-18 | | Release date: | 2007-01-23 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of cholesteryl ester transfer protein reveals a long tunnel and four bound lipid molecules.
Nat.Struct.Mol.Biol., 14, 2007
|
|
1NMT
 
 | |
4EWS
 
 | |
1QG6
 
 | |
3KWB
 
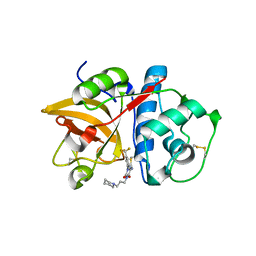 | | Structure of CatK covalently bound to a dioxo-triazine inhibitor | | Descriptor: | 3,5-dioxo-4-(3-piperidin-1-ylpropyl)-2-[3-(trifluoromethyl)phenyl]-2,3,4,5-tetrahydro-1,2,4-triazine-6-carbonitrile, Cathepsin K | | Authors: | Uitdehaag, J.C.M, van Zeeland, M. | | Deposit date: | 2009-12-01 | | Release date: | 2010-04-07 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Dioxo-triazines as a novel series of cathepsin K inhibitors
Bioorg.Med.Chem.Lett., 20, 2010
|
|
3KW9
 
 | |
3KWZ
 
 | |
3KX1
 
 | |
1D8A
 
 | | E. COLI ENOYL REDUCTASE/NAD+/TRICLOSAN COMPLEX | | Descriptor: | ENOYL-[ACYL-CARRIER-PROTEIN] REDUCTASE, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, TRICLOSAN | | Authors: | Levy, C.W, Roujeinikova, A, Sedelnikova, S, Baker, P.J, Stuitje, A.R, Slabas, A.R, Rice, D.W, Rafferty, J.B. | | Deposit date: | 1999-10-21 | | Release date: | 1999-10-28 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Molecular basis of triclosan activity.
Nature, 398, 1999
|
|
4F2A
 
 | | Crystal structure of cholestryl esters transfer protein in complex with inhibitors | | Descriptor: | (2R)-3-{[4-(4-chloro-3-ethylphenoxy)pyrimidin-2-yl][3-(1,1,2,2-tetrafluoroethoxy)benzyl]amino}-1,1,1-trifluoropropan-2-ol, 1,2-DIOLEOYL-SN-GLYCERO-3-PHOSPHOCHOLINE, CHLORIDE ION, ... | | Authors: | Liu, S, Qiu, X. | | Deposit date: | 2012-05-07 | | Release date: | 2012-09-05 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (3.11 Å) | | Cite: | Crystal structures of cholesteryl ester transfer protein in complex with inhibitors.
J.Biol.Chem., 287, 2012
|
|
3FZP
 
 | | Crystal structure of PYK2 complexed with ATPgS | | Descriptor: | PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER, Protein tyrosine kinase 2 beta, SULFATE ION | | Authors: | Han, S. | | Deposit date: | 2009-01-26 | | Release date: | 2009-03-31 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural characterization of proline-rich tyrosine kinase 2 (PYK2) reveals a unique (DFG-out) conformation and enables inhibitor design.
J.Biol.Chem., 284, 2009
|
|
3FZT
 
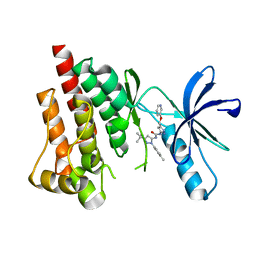 | | Crystal structure of PYK2 complexed with PF-4618433 | | Descriptor: | 1-[5-tert-butyl-2-(4-methylphenyl)-1,2-dihydro-3H-pyrazol-3-ylidene]-3-{3-[(pyridin-3-yloxy)methyl]-1H-pyrazol-5-yl}urea, Protein tyrosine kinase 2 beta | | Authors: | Han, S. | | Deposit date: | 2009-01-26 | | Release date: | 2009-03-31 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural characterization of proline-rich tyrosine kinase 2 (PYK2) reveals a unique (DFG-out) conformation and enables inhibitor design.
J.Biol.Chem., 284, 2009
|
|
3FZO
 
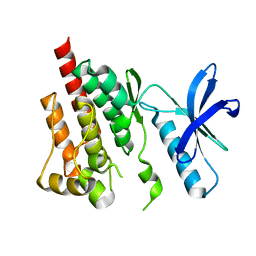 | | Crystal Structure of PYK2-Apo, Proline-rich Tyrosine Kinase | | Descriptor: | Protein tyrosine kinase 2 beta | | Authors: | Han, S. | | Deposit date: | 2009-01-26 | | Release date: | 2009-03-31 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural characterization of proline-rich tyrosine kinase 2 (PYK2) reveals a unique (DFG-out) conformation and enables inhibitor design.
J.Biol.Chem., 284, 2009
|
|
3FZR
 
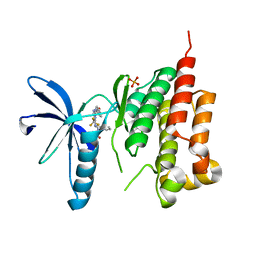 | | Crystal structure of PYK2 complexed with PF-431396 | | Descriptor: | N-methyl-N-{2-[({2-[(2-oxo-2,3-dihydro-1H-indol-5-yl)amino]-5-(trifluoromethyl)pyrimidin-4-yl}amino)methyl]phenyl}methanesulfonamide, PHOSPHATE ION, Protein tyrosine kinase 2 beta | | Authors: | Han, S. | | Deposit date: | 2009-01-26 | | Release date: | 2009-03-31 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural characterization of proline-rich tyrosine kinase 2 (PYK2) reveals a unique (DFG-out) conformation and enables inhibitor design.
J.Biol.Chem., 284, 2009
|
|
