6I2V
 
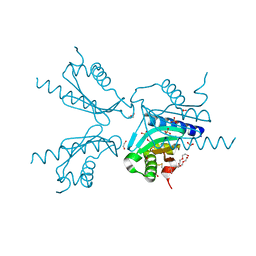 | | Pilotin from Vibrio vulnificus type 2 secretion system, EpsS. | | Descriptor: | 1,2-ETHANEDIOL, PENTAETHYLENE GLYCOL, SULFATE ION, ... | | Authors: | Howard, S.P, Estrozi, L, Bertrand, Q, Contreras-Martel, C, Strozen, T, Job, V, Martins, A, Fenel, D, Schoehn, G, Dessen, A. | | Deposit date: | 2018-11-02 | | Release date: | 2019-04-10 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structure and assembly of pilotin-dependent and -independent secretins of the type II secretion system.
Plos Pathog., 15, 2019
|
|
1YN9
 
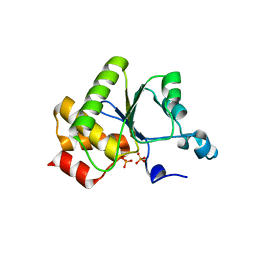 | | Crystal structure of baculovirus RNA 5'-phosphatase complexed with phosphate | | Descriptor: | PHOSPHATE ION, polynucleotide 5'-phosphatase | | Authors: | Changela, A, Martins, A, Shuman, S, Mondragon, A. | | Deposit date: | 2005-01-24 | | Release date: | 2005-02-22 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structure of baculovirus RNA triphosphatase complexed with phosphate
J.Biol.Chem., 280, 2005
|
|
4U39
 
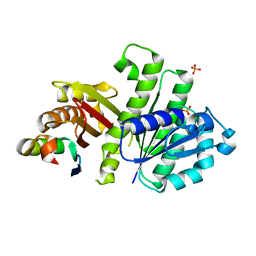 | | Crystal Structure of FtsZ:MciZ Complex from Bacillus subtilis | | Descriptor: | Cell division factor, Cell division protein FtsZ, PHOSPHATE ION | | Authors: | Bisson-Filho, A.W, Discola, K.F, Castellen, P, Blasios, V, Martins, A, Sforca, M.L, Garcia, W, Zeri, A.C, Erickson, H.P, Dessen, A, Gueiros-Filho, F.J. | | Deposit date: | 2014-07-19 | | Release date: | 2015-03-18 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.194 Å) | | Cite: | Crystal Structure of FtsZ:MciZ Complex from Bacillus subtilis
To be Published
|
|
1I9S
 
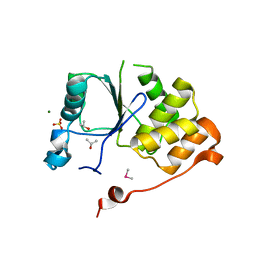 | | CRYSTAL STRUCTURE OF THE RNA TRIPHOSPHATASE DOMAIN OF MOUSE MRNA CAPPING ENZYME | | Descriptor: | CACODYLATE ION, ISOPROPYL ALCOHOL, MAGNESIUM ION, ... | | Authors: | Changela, A, Ho, C.K, Martins, A, Shuman, S, Mondragon, A. | | Deposit date: | 2001-03-20 | | Release date: | 2001-05-23 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structure and mechanism of the RNA triphosphatase component of mammalian mRNA capping enzyme.
EMBO J., 20, 2001
|
|
1I9T
 
 | | CRYSTAL STRUCTURE OF THE OXIDIZED RNA TRIPHOSPHATASE DOMAIN OF MOUSE MRNA CAPPING ENZYME | | Descriptor: | CACODYLATE ION, ISOPROPYL ALCOHOL, MAGNESIUM ION, ... | | Authors: | Changela, A, Ho, C.K, Martins, A, Shuman, S, Mondragon, A. | | Deposit date: | 2001-03-20 | | Release date: | 2001-05-23 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure and mechanism of the RNA triphosphatase component of mammalian mRNA capping enzyme.
EMBO J., 20, 2001
|
|
5LP4
 
 | | Penicillin-Binding Protein (PBP2) from Helicobacter pylori | | Descriptor: | Penicillin-binding protein 2 (Pbp2), SULFATE ION | | Authors: | Contreras-Martel, C, Martins, A, Ecobichon, C, Maragno, D.M, Mattei, P.J, El Ghachi, M, Boneca, I.G, Dessen, A. | | Deposit date: | 2016-08-11 | | Release date: | 2017-08-23 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.03 Å) | | Cite: | Molecular architecture of the PBP2-MreC core bacterial cell wall synthesis complex.
Nat Commun, 8, 2017
|
|
5LP5
 
 | | Complex between Penicillin-Binding Protein (PBP2) and MreC from Helicobacter pylori | | Descriptor: | Penicillin-binding protein 2 (Pbp2), Rod shape-determining protein (MreC) | | Authors: | Contreras-Martel, C, Martins, A, Ecobichon, C, Maragno, D.M, Mattei, P.J, El Ghachi, M, Hicham, S, Hardouin, P, Boneca, I.G, Dessen, A. | | Deposit date: | 2016-08-11 | | Release date: | 2017-08-23 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.74 Å) | | Cite: | Molecular architecture of the PBP2-MreC core bacterial cell wall synthesis complex.
Nat Commun, 8, 2017
|
|
2Y2G
 
 | | PENICILLIN-BINDING PROTEIN 1B (PBP-1B) IN COMPLEX WITH AN ALKYL BORONATE (A01) | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, PENICILLIN-BINDING PROTEIN 1B, ... | | Authors: | Contreras-Martel, C, Amoroso, A, Woon, E.C, Zervosen, A, Inglis, S, Martins, A, Verlaine, O, Rydzik, A, Job, V, Luxen, A, Joris, B, Schofield, C.J, Dessen, A. | | Deposit date: | 2010-12-15 | | Release date: | 2011-08-03 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structure-Guided Design of Cell Wall Biosynthesis Inhibitors that Overcome Beta-Lactam Resistance in Staphylococcus Aureus (Mrsa).
Acs Chem.Biol., 6, 2011
|
|
2Y2N
 
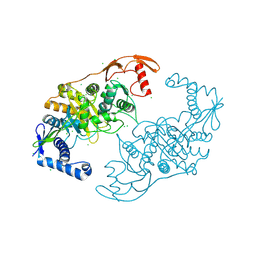 | | PENICILLIN-BINDING PROTEIN 1B (PBP-1B) IN COMPLEX WITH AN ALKYL BORONATE (E07) | | Descriptor: | CHLORIDE ION, PENICILLIN-BINDING PROTEIN 1B, SODIUM ION, ... | | Authors: | Contreras-Martel, C, Amoroso, A, Woon, E.C, Zervosen, A, Inglis, S, Martins, A, Verlaine, O, Rydzik, A, Job, V, Luxen, A, Joris, B, Schofield, C.J, Dessen, A. | | Deposit date: | 2010-12-15 | | Release date: | 2011-08-03 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Structure-Guided Design of Cell Wall Biosynthesis Inhibitors that Overcome Beta-Lactam Resistance in Staphylococcus Aureus (Mrsa).
Acs Chem.Biol., 6, 2011
|
|
2Y2M
 
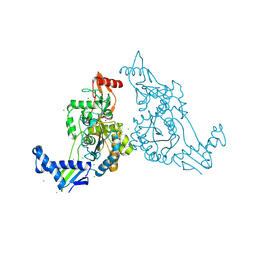 | | PENICILLIN-BINDING PROTEIN 1B (PBP-1B) IN COMPLEX WITH AN ALKYL BORONATE (E08) | | Descriptor: | CHLORIDE ION, PENICILLIN-BINDING PROTEIN 1B, SODIUM ION, ... | | Authors: | Contreras-Martel, C, Amoroso, A, Woon, E.C, Zervosen, A, Inglis, S, Martins, A, Verlaine, O, Rydzik, A, Job, V, Luxen, A, Joris, B, Schofield, C.J, Dessen, A. | | Deposit date: | 2010-12-15 | | Release date: | 2011-08-03 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Structure-Guided Design of Cell Wall Biosynthesis Inhibitors that Overcome Beta-Lactam Resistance in Staphylococcus Aureus (Mrsa).
Acs Chem.Biol., 6, 2011
|
|
2Y2J
 
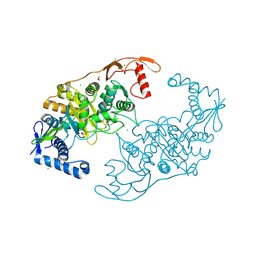 | | PENICILLIN-BINDING PROTEIN 1B (PBP-1B) IN COMPLEX WITH AN ALKYL BORONATE (ZA4) | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, PENICILLIN-BINDING PROTEIN 1B, ... | | Authors: | Contreras-Martel, C, Amoroso, A, Woon, E.C, Zervosen, A, Inglis, S, Martins, A, Verlaine, O, Rydzik, A, Job, V, Luxen, A, Joris, B, Schofield, C.J, Dessen, A. | | Deposit date: | 2010-12-15 | | Release date: | 2011-08-03 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Structure-Guided Design of Cell Wall Biosynthesis Inhibitors that Overcome Beta-Lactam Resistance in Staphylococcus Aureus (Mrsa).
Acs Chem.Biol, 6, 2011
|
|
2Y2K
 
 | | PENICILLIN-BINDING PROTEIN 1B (PBP-1B) IN COMPLEX WITH AN ALKYL BORONATE (ZA5) | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, PENICILLIN-BINDING PROTEIN 1B, ... | | Authors: | Contreras-Martel, C, Amoroso, A, Woon, E, Zervosen, A, Inglis, S, Martins, A, Verlaine, O, Rydzik, A, Job, V, Luxen, A, Joris, B, Schofield, C, Dessen, A. | | Deposit date: | 2010-12-15 | | Release date: | 2011-08-03 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Structure-Guided Design of Cell Wall Biosynthesis Inhibitors that Overcome Beta-Lactam Resistance in Staphylococcus Aureus (Mrsa).
Acs Chem.Biol, 6, 2011
|
|
2Y2O
 
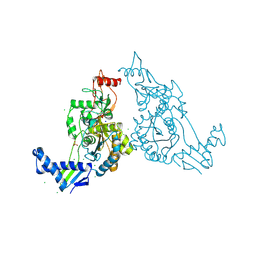 | | PENICILLIN-BINDING PROTEIN 1B (PBP-1B) IN COMPLEX WITH AN ALKYL BORONATE (EO9) | | Descriptor: | CHLORIDE ION, PENICILLIN-BINDING PROTEIN 1B, SODIUM ION, ... | | Authors: | Contreras-Martel, C, Amoroso, A, Woon, E.C, Zervosen, A, Inglis, S, Martins, A, Verlaine, O, Rydzik, A, Job, V, Luxen, A, Joris, B, Schofield, C.J, Dessen, A. | | Deposit date: | 2010-12-15 | | Release date: | 2011-08-03 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Structure-Guided Design of Cell Wall Biosynthesis Inhibitors that Overcome Beta-Lactam Resistance in Staphylococcus Aureus (Mrsa).
Acs Chem.Biol., 6, 2011
|
|
2Y2P
 
 | | Penicillin-binding protein 1b (pbp-1b) in complex with an alkyl boronate (z10) | | Descriptor: | CHLORIDE ION, PENICILLIN-BINDING PROTEIN 1B, SODIUM ION, ... | | Authors: | Contreras-Martel, C, Amoroso, A, Woon, E.C, Zervosen, A, Inglis, S, Martins, A, Verlaine, O, Rydzik, A, Job, V, Luxen, A, Joris, B, Schofield, C.J, Dessen, A. | | Deposit date: | 2010-12-15 | | Release date: | 2011-08-03 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Structure-Guided Design of Cell Wall Biosynthesis Inhibitors that Overcome Beta-Lactam Resistance in Staphylococcus Aureus (Mrsa).
Acs Chem.Biol., 6, 2011
|
|
2Y2I
 
 | | PENICILLIN-BINDING PROTEIN 1B (PBP-1B) IN COMPLEX WITH AN ALKYL BORONATE (ZA3) | | Descriptor: | CHLORIDE ION, PENICILLIN-BINDING PROTEIN 1B, SODIUM ION, ... | | Authors: | Contreras-Martel, C, Amoroso, A, Woon, E.C, Zervosen, A, Inglis, S, Martins, A, Verlaine, O, Rydzik, A, Job, V, Luxen, A, Joris, B, Schofield, C.J, Dessen, A. | | Deposit date: | 2010-12-15 | | Release date: | 2011-08-03 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Structure-Guided Design of Cell Wall Biosynthesis Inhibitors that Overcome Beta-Lactam Resistance in Staphylococcus Aureus (Mrsa).
Acs Chem.Biol., 6, 2011
|
|
2Y2L
 
 | | PENICILLIN-BINDING PROTEIN 1B (PBP-1B) IN COMPLEX WITH AN ALKYL BORONATE (E06) | | Descriptor: | CHLORIDE ION, PENICILLIN-BINDING PROTEIN 1B, SODIUM ION, ... | | Authors: | Contreras-Martel, C, Amoroso, A, Woon, E.C, Zervosen, A, Inglis, S, Martins, A, Verlaine, O, Rydzik, A, Job, V, Luxen, A, Joris, B, Schofield, C.J, Dessen, A. | | Deposit date: | 2010-12-15 | | Release date: | 2011-08-03 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Structure-Guided Design of Cell Wall Biosynthesis Inhibitors that Overcome Beta-Lactam Resistance in Staphylococcus Aureus (Mrsa).
Acs Chem.Biol., 6, 2011
|
|
2Y2H
 
 | | PENICILLIN-BINDING PROTEIN 1B (PBP-1B) IN COMPLEX WITH AN ALKYL BORONATE (ZA2) | | Descriptor: | CHLORIDE ION, PENICILLIN-BINDING PROTEIN 1B, SODIUM ION, ... | | Authors: | Contreras-Martel, C, Amoroso, A, Woon, E.C, Zervosen, A, Inglis, S, Martins, A, Verlaine, O, Rydzik, A, Job, V, Luxen, A, Joris, B, Schofield, C.J, Dessen, A. | | Deposit date: | 2010-12-15 | | Release date: | 2011-08-03 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Structure-Guided Design of Cell Wall Biosynthesis Inhibitors that Overcome Beta-Lactam Resistance in Staphylococcus Aureus (Mrsa).
Acs Chem.Biol., 6, 2011
|
|
2Y2Q
 
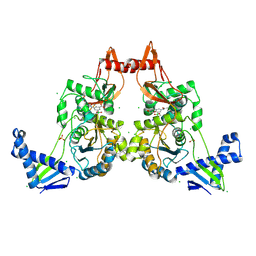 | | PENICILLIN-BINDING PROTEIN 1B (PBP-1B) IN COMPLEX WITH AN ALKYL BORONATE (Z06) | | Descriptor: | (3-acetamido-5-carboxy-phenyl)-trihydroxy-boron, 1,2-ETHANEDIOL, CHLORIDE ION, ... | | Authors: | Contreras-Martel, C, Amoroso, A, Woon, E.C, Zervosen, A, Inglis, S, Martins, A, Verlaine, O, Rydzik, A, Job, V, Luxen, A, Joris, B, Schofield, C.J, Dessen, A. | | Deposit date: | 2010-12-15 | | Release date: | 2011-08-03 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structure-Guided Design of Cell Wall Biosynthesis Inhibitors that Overcome Beta-Lactam Resistance in Staphylococcus Aureus (Mrsa).
Acs Chem.Biol., 6, 2011
|
|
5DTY
 
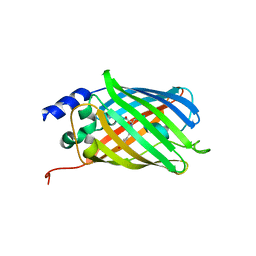 | |
2WAD
 
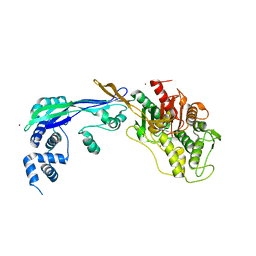 | | PENICILLIN-BINDING PROTEIN 2B (PBP-2B) FROM STREPTOCOCCUS PNEUMONIAE (STRAIN 5204) | | Descriptor: | PENICILLIN-BINDING PROTEIN 2B, ZINC ION | | Authors: | Contreras-Martel, C, Dahout-Gonzalez, C, Dos-Santos-Martins, A, Kotnik, M, Dessen, A. | | Deposit date: | 2009-02-05 | | Release date: | 2009-02-24 | | Last modified: | 2014-02-19 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | Pbp Active Site Flexibility as the Key Mechanism for Beta-Lactam Resistance in Pneumococci
J.Mol.Biol., 387, 2009
|
|
2WAF
 
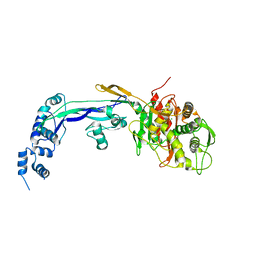 | | PENICILLIN-BINDING PROTEIN 2B (PBP-2B) FROM STREPTOCOCCUS PNEUMONIAE (STRAIN R6) | | Descriptor: | CHLORIDE ION, PENICILLIN-BINDING PROTEIN 2B | | Authors: | Contreras-Martel, C, Dahout-Gonzalez, C, Dos-Santos-Martins, A, Kotnik, M, Dessen, A. | | Deposit date: | 2009-02-05 | | Release date: | 2009-02-24 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (3.29 Å) | | Cite: | Pbp Active Site Flexibility as the Key Mechanism for Beta-Lactam Resistance in Pneumococci
J.Mol.Biol., 387, 2009
|
|
2WAE
 
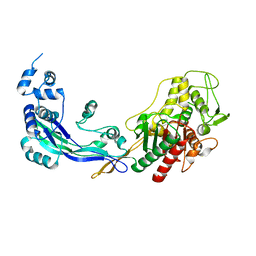 | | PENICILLIN-BINDING PROTEIN 2B (PBP-2B) FROM STREPTOCOCCUS PNEUMONIAE (STRAIN 5204) | | Descriptor: | PENICILLIN-BINDING PROTEIN 2B, ZINC ION | | Authors: | Contreras-Martel, C, Dahout-Gonzalez, C, Dos-Santos-Martins, A, Kotnik, M, Dessen, A. | | Deposit date: | 2009-02-05 | | Release date: | 2009-02-24 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | Pbp Active Site Flexibility as the Key Mechanism for Beta-Lactam Resistance in Pneumococci
J.Mol.Biol., 387, 2009
|
|
7ZUH
 
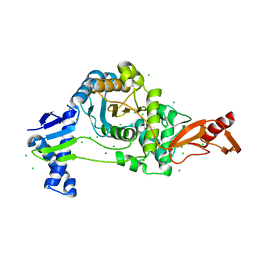 | | PENICILLIN-BINDING PROTEIN 1B (PBP-1B) Streptococcus pneumoniae R6 | | Descriptor: | CHLORIDE ION, MAGNESIUM ION, Penicillin-binding protein 1b | | Authors: | Flanders, P.L, Contreras-Martel, C, Martins, A, Brown, N.W, Shirley, J.D, Nauta, K.M, Dessen, A, Carlson, E.E, Ambrose, E.A. | | Deposit date: | 2022-05-12 | | Release date: | 2022-11-02 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.467 Å) | | Cite: | Combined Structural Analysis and Molecular Dynamics Reveal Penicillin-Binding Protein Inhibition Mode with beta-Lactones.
Acs Chem.Biol., 17, 2022
|
|
7ZUK
 
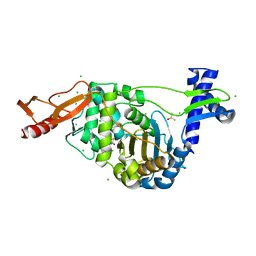 | | PENICILLIN-BINDING PROTEIN 1B (PBP-1B) in complex with lactone 7Az - Streptococcus pneumoniae R6 | | Descriptor: | 6-azido-N-[(2S)-1-oxidanylidene-1-[[(2S,3R)-3-oxidanyl-1-oxidanylidene-butan-2-yl]amino]-3-phenyl-propan-2-yl]hexanamide, CHLORIDE ION, DIMETHYL SULFOXIDE, ... | | Authors: | Flanders, P.L, Contreras-Martel, C, Martins, A, Brown, N.W, Shirley, J.D, Nauta, K.M, Dessen, A, Carlson, E.E, Ambrose, E.A. | | Deposit date: | 2022-05-12 | | Release date: | 2022-11-02 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.631 Å) | | Cite: | Combined Structural Analysis and Molecular Dynamics Reveal Penicillin-Binding Protein Inhibition Mode with beta-Lactones.
Acs Chem.Biol., 17, 2022
|
|
7ZUJ
 
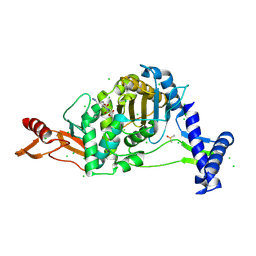 | | PENICILLIN-BINDING PROTEIN 1B (PBP-1B) in complex with lactone 6Az - Streptococcus pneumoniae R6 | | Descriptor: | 6-azido-N-[(2S)-1-oxidanylidene-1-[[(2S,3R)-3-oxidanyl-1-oxidanylidene-butan-2-yl]amino]propan-2-yl]hexanamide, CHLORIDE ION, DIMETHYL SULFOXIDE, ... | | Authors: | Flanders, P.L, Contreras-Martel, C, Martins, A, Brown, N.W, Shirley, J.D, Nauta, K.M, Dessen, A, Carlson, E.E, Ambrose, E.A. | | Deposit date: | 2022-05-12 | | Release date: | 2022-11-02 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Combined Structural Analysis and Molecular Dynamics Reveal Penicillin-Binding Protein Inhibition Mode with beta-Lactones.
Acs Chem.Biol., 17, 2022
|
|
