4JSU
 
 | | Yeast 20S proteasome in complex with the dimerized linear mimetic of TMC-95A - yCP:3a | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Probable proteasome subunit alpha type-7, Proteasome subunit alpha type-1, ... | | Authors: | Desvergne, A, Genin, E, Marechal, X, Gallastegui, N, Dufau, L, Richy, N, Groll, M, Vidal, J, Reboud-Ravaux, M. | | Deposit date: | 2013-03-22 | | Release date: | 2013-05-01 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Dimerized linear mimics of a natural cyclopeptide (TMC-95A) are potent noncovalent inhibitors of the eukaryotic 20S proteasome
J.Med.Chem., 56, 2013
|
|
4JSQ
 
 | | Yeast 20S proteasome in complex with the dimerized linear mimetic of TMC-95A - yCP:4e | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Probable proteasome subunit alpha type-7, Proteasome subunit alpha type-1, ... | | Authors: | Desvergne, A, Genin, E, Marechal, X, Gallastegui, N, Dufau, L, Richy, N, Groll, M, Vidal, J, Reboud-Ravaux, M. | | Deposit date: | 2013-03-22 | | Release date: | 2013-05-01 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Dimerized linear mimics of a natural cyclopeptide (TMC-95A) are potent noncovalent inhibitors of the eukaryotic 20S proteasome
J.Med.Chem., 56, 2013
|
|
4JT0
 
 | | Yeast 20S proteasome in complex with the dimerized linear mimetic of TMC-95A - yCP:4a | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Probable proteasome subunit alpha type-7, Proteasome subunit alpha type-1, ... | | Authors: | Desvergne, A, Genin, E, Marechal, X, Gallastegui, N, Dufau, L, Richy, N, Groll, M, Vidal, J, Reboud-Ravaux, M. | | Deposit date: | 2013-03-22 | | Release date: | 2013-05-01 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Dimerized linear mimics of a natural cyclopeptide (TMC-95A) are potent noncovalent inhibitors of the eukaryotic 20S proteasome.
J.Med.Chem., 56, 2013
|
|
3NZW
 
 | | Crystal structure of the yeast 20S proteasome in complex with 2b | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Proteasome component C1, Proteasome component C11, ... | | Authors: | Groll, M, Gallastegui, N, Marechal, X, Le Ravalec, V, Basse, N, Richy, N, Genin, E, Huber, R, Moroder, M, Vidal, V, Reboud-Ravaux, M. | | Deposit date: | 2010-07-17 | | Release date: | 2011-02-16 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | 20S proteasome inhibition: designing noncovalent linear peptide mimics of the natural product TMC-95A.
Chemmedchem, 5, 2010
|
|
3NZJ
 
 | | Crystal structure of yeast 20S proteasome in complex with ligand 2a | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Proteasome component C1, Proteasome component C11, ... | | Authors: | Groll, M, Gallastegui, N, Marechal, X, Le Ravalec, V, Basse, N, Richy, N, Genin, E, Huber, R, Moroder, M, Vidal, V, Reboud-Ravaux, M. | | Deposit date: | 2010-07-16 | | Release date: | 2011-02-16 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | 20S proteasome inhibition: designing noncovalent linear peptide mimics of the natural product TMC-95A.
Chemmedchem, 5, 2010
|
|
3NZX
 
 | | Crystal structure of the yeast 20S proteasome in complex with ligand 2c | | Descriptor: | Proteasome component C1, Proteasome component C11, Proteasome component C5, ... | | Authors: | Groll, M, Gallastegui, N, Marechal, X, Le Ravalec, V, Basse, N, Richy, N, Genin, E, Huber, R, Moroder, M, Vidal, V, Reboud-Ravaux, M. | | Deposit date: | 2010-07-17 | | Release date: | 2011-02-16 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | 20S proteasome inhibition: designing noncovalent linear peptide mimics of the natural product TMC-95A.
Chemmedchem, 5, 2010
|
|
2YPU
 
 | | human insulin degrading enzyme E111Q in complex with inhibitor compound 41367 | | Descriptor: | 2-[[2-[[(2S)-3-(3H-IMIDAZOL-4-YL)-1-METHOXY-1-OXO-PROPAN-2-YL]AMINO]-2-OXO-ETHYL]-(PHENYLMETHYL)AMINO]ETHANOIC ACID, INSULIN-DEGRADING ENZYME, ZINC ION | | Authors: | Guo, Q, Deprez-Poulain, R, Deprez, B, Tang, W.-J. | | Deposit date: | 2012-11-01 | | Release date: | 2012-11-28 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Imidazole-Derived 2-[N-Carbamoylmethyl-Alkylamino]Acetic Acids,Substrate-Dependent Modulators of Insulin-Degrading Enzyme in Amyloid-Beta Hydrolysis
Eur J Med Chem, 79C, 2014
|
|
4QIA
 
 | | Crystal structure of human insulin degrading enzyme (ide) in complex with inhibitor N-benzyl-N-(carboxymethyl)glycyl-L-histidine | | Descriptor: | Insulin-degrading enzyme, N-benzyl-N-(carboxymethyl)glycyl-L-histidine, ZINC ION | | Authors: | Guo, Q, Deprez-Poulain, R, Deprez, B, Tang, W.J. | | Deposit date: | 2014-05-30 | | Release date: | 2015-05-13 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.202 Å) | | Cite: | Structure-activity relationships of imidazole-derived 2-[N-carbamoylmethyl-alkylamino]acetic acids, dual binders of human insulin-degrading enzyme.
Eur.J.Med.Chem., 90, 2015
|
|
4RE9
 
 | | Crystal structure of human insulin degrading enzyme (IDE) in complex with compound 71290 | | Descriptor: | 1,2-ETHANEDIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, 4-fluoro-N-({1-[(2R)-4-(hydroxyamino)-1-(naphthalen-2-yl)-4-oxobutan-2-yl]-1H-1,2,3-triazol-5-yl}methyl)benzamide, ... | | Authors: | Liang, W.G, Deprez, R, Deprez, B, Tang, W.J. | | Deposit date: | 2014-09-22 | | Release date: | 2015-09-30 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.908 Å) | | Cite: | Catalytic site inhibition of insulin-degrading enzyme by a small molecule induces glucose intolerance in mice.
Nat Commun, 6, 2015
|
|
4GSF
 
 | | The structure analysis of cysteine free insulin degrading enzyme (ide) with (s)-2-{2-[carboxymethyl-(3-phenyl-propionyl)-amino]-acetylamino}-3-(3h-imidazol-4-yl)-propionic acid methyl ester | | Descriptor: | Insulin-degrading enzyme, ZINC ION, methyl N-(carboxymethyl)-N-(3-phenylpropanoyl)glycyl-D-histidinate | | Authors: | Guo, Q, Deprez-Poulain, R, Deprez, B, Tang, W.J. | | Deposit date: | 2012-08-27 | | Release date: | 2013-08-28 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structure-activity relationships of imidazole-derived 2-[N-carbamoylmethyl-alkylamino]acetic acids, dual binders of human insulin-degrading enzyme.
Eur.J.Med.Chem., 90, 2015
|
|
4NXO
 
 | | Crystal Structure of Insulin Degrading Enzyme in complex with BDM44768 | | Descriptor: | 1,2-ETHANEDIOL, 1,4-DIETHYLENE DIOXIDE, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ... | | Authors: | Liang, W.G, Deprez, R, Deprez, B, Tang, W. | | Deposit date: | 2013-12-09 | | Release date: | 2015-10-07 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.73 Å) | | Cite: | Catalytic site inhibition of insulin-degrading enzyme by a small molecule induces glucose intolerance in mice.
Nat Commun, 6, 2015
|
|
4IFH
 
 | | Crystal structure of human insulin degrading enzyme (IDE) in complex with compound BDM44619 | | Descriptor: | Insulin-degrading enzyme, N-({1-[(2R)-4-(hydroxyamino)-1-(naphthalen-2-yl)-4-oxobutan-2-yl]-1H-1,2,3-triazol-4-yl}methyl)-4-methylbenzamide, ZINC ION | | Authors: | Liang, W.G, Guo, Q, Deprez, R, Deprez, B, Tang, W. | | Deposit date: | 2012-12-14 | | Release date: | 2013-12-18 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.286 Å) | | Cite: | Catalytic site inhibition of insulin-degrading enzyme by a small molecule induces glucose intolerance in mice.
Nat Commun, 6, 2015
|
|
4DWK
 
 | | Structure of cystein free insulin degrading enzyme with compound bdm41671 ((s)-2-{2-[carboxymethyl-(3-phenyl-propyl)-amino]-acetylamino}-3-(1h-imidazol-4-yl)-propionic acid methyl ester) | | Descriptor: | Insulin-degrading enzyme, ZINC ION, methyl N-(carboxymethyl)-N-(3-phenylpropyl)glycyl-L-histidinate | | Authors: | Guo, Q, Deprez-Poulain, R, Deprez, B, Tang, W.J. | | Deposit date: | 2012-02-24 | | Release date: | 2013-04-03 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Imidazole-derived 2-[N-carbamoylmethyl-alkylamino]acetic acids, substrate-dependent modulators of insulin-degrading enzyme in amyloid-beta hydrolysis.
Eur.J.Med.Chem., 79, 2014
|
|
4DTT
 
 | | Crystal structure of human insulin degrading enzyme (ide) in complex with compund 41367 | | Descriptor: | 2-[[2-[[(2S)-3-(3H-IMIDAZOL-4-YL)-1-METHOXY-1-OXO-PROPAN-2-YL]AMINO]-2-OXO-ETHYL]-(PHENYLMETHYL)AMINO]ETHANOIC ACID, Insulin-degrading enzyme, ZINC ION | | Authors: | Guo, Q, Deprez-Poulain, R, Deprez, B, Tang, W.J. | | Deposit date: | 2012-02-21 | | Release date: | 2013-02-27 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3.22 Å) | | Cite: | Imidazole-derived 2-[N-carbamoylmethyl-alkylamino]acetic acids, substrate-dependent modulators of insulin-degrading enzyme in amyloid-beta hydrolysis.
Eur.J.Med.Chem., 79, 2014
|
|
4GS8
 
 | | Structure analysis of cysteine free insulin degrading enzyme (ide) with compound bdm43079 [{[(s)-2-(1h-imidazol-4-yl)-1-methylcarbamoyl-ethylcarbamoyl]-methyl}-(3-phenyl-propyl)-amino]-acetic acid | | Descriptor: | Insulin-degrading enzyme, N-(carboxymethyl)-N-(3-phenylpropyl)glycyl-N-methyl-L-histidinamide, ZINC ION | | Authors: | Guo, Q, Deprez-Poulain, R, Deprez, B, Tang, W.J. | | Deposit date: | 2012-08-27 | | Release date: | 2013-08-28 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.99 Å) | | Cite: | Imidazole-derived 2-[N-carbamoylmethyl-alkylamino]acetic acids, substrate-dependent modulators of insulin-degrading enzyme in amyloid-beta hydrolysis.
Eur.J.Med.Chem., 79, 2014
|
|
4GSC
 
 | | Structure analysis of insulin degrading enzyme with compound bdm41559 ((s)-2-[2-(carboxymethyl-phenethyl-amino)-acetylamino]-3-(1h-imidazol-4-yl)-propionic acid methyl ester) | | Descriptor: | Insulin-degrading enzyme, ZINC ION, methyl N-(carboxymethyl)-N-(2-phenylethyl)glycyl-L-histidinate | | Authors: | Guo, Q, Deprez-Poulain, R, Deprez, B, Tang, W.J. | | Deposit date: | 2012-08-27 | | Release date: | 2013-08-28 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.81 Å) | | Cite: | Imidazole-derived 2-[N-carbamoylmethyl-alkylamino]acetic acids, substrate-dependent modulators of insulin-degrading enzyme in amyloid-beta hydrolysis.
Eur.J.Med.Chem., 79, 2014
|
|
3QZ2
 
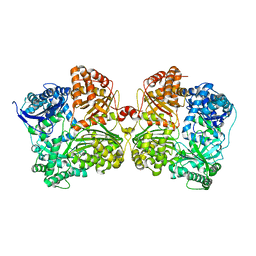 | | The structure of cysteine-free human insulin degrading enzyme | | Descriptor: | Insulin-degrading enzyme, ZINC ION | | Authors: | Guo, Q, Tang, W.J. | | Deposit date: | 2011-03-04 | | Release date: | 2012-01-25 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Imidazole-derived 2-[N-carbamoylmethyl-alkylamino]acetic acids, substrate-dependent modulators of insulin-degrading enzyme in amyloid-beta hydrolysis.
Eur.J.Med.Chem., 79, 2014
|
|
