2VZ8
 
 | |
2VZ9
 
 | |
4QKY
 
 | | Crystal Structure Analysis of the Membrane Transporter FhaC | | Descriptor: | Filamentous hemagglutinin transporter protein FhaC, PHOSPHATE ION | | Authors: | Maier, T, Clantin, B, Gruss, F, Dewitte, F, Delattre, A.S, Jacob-Dubuisson, F, Hiller, S, Villeret, V. | | Deposit date: | 2014-06-10 | | Release date: | 2014-10-22 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Conserved Omp85 lid-lock structure and substrate recognition in FhaC
Nat Commun, 6, 2015
|
|
4QL0
 
 | | Crystal Structure Analysis of the Membrane Transporter FhaC (double mutant V169T, I176N) | | Descriptor: | DI(HYDROXYETHYL)ETHER, Filamentous hemagglutinin transporter protein FhaC, HEXAETHYLENE GLYCOL, ... | | Authors: | Maier, T, Clantin, B, Gruss, F, Dewitte, F, Delattre, A.S, Jacob-Dubuisson, F, Hiller, S, Villeret, V. | | Deposit date: | 2014-06-10 | | Release date: | 2015-06-17 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Conserved Omp85 lid-lock structure and substrate recognition in FhaC
Nat Commun, 6, 2015
|
|
1O7A
 
 | | Human beta-Hexosaminidase B | | Descriptor: | 1,2-ETHANEDIOL, 2-(acetylamido)-2-deoxy-D-glucono-1,5-lactone, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Maier, T, Strater, N, Schuette, C, Klingenstein, R, Sandhoff, K, Saenger, W. | | Deposit date: | 2002-10-29 | | Release date: | 2003-10-23 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | The X-Ray Crystal Structure of Human Beta-Hexosaminidase B Provides New Insights Into Sandhoff Disease
J.Mol.Biol., 328, 2003
|
|
2BJ6
 
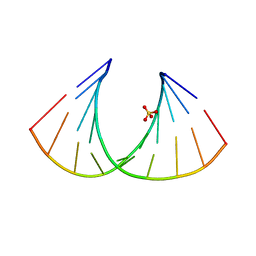 | | Crystal Structure of a decameric HNA-RNA hybrid | | Descriptor: | 5'-R(*GP*GP*CP*AP*UP*UP*AP*CP*GP*GP)-3', SULFATE ION, SYNTHETIC HNA | | Authors: | Maier, T, Przylas, I, Straeter, N, Herdewijn, P, Saenger, W. | | Deposit date: | 2005-01-30 | | Release date: | 2005-03-09 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Reinforced Hna Backbone Hydration in the Crystal Structure of a Decameric Hna/RNA Hybrid
J.Am.Chem.Soc., 127, 2005
|
|
2CF2
 
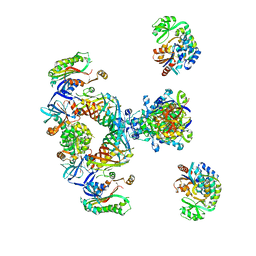 | | Architecture of mammalian fatty acid synthase | | Descriptor: | FATTY ACID SYNTHASE, DH DOMAIN, ER DOMAIN, ... | | Authors: | Maier, T, Jenni, S, Ban, N. | | Deposit date: | 2006-02-14 | | Release date: | 2006-03-07 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (4.3 Å) | | Cite: | Architecture of Mammalian Fatty Acid Synthase at 4.5 A Resolution.
Science, 311, 2006
|
|
2R1Q
 
 | |
2RB3
 
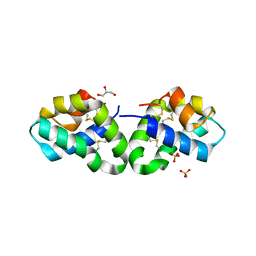 | | Crystal Structure of Human Saposin D | | Descriptor: | GLYCEROL, Proactivator polypeptide, SULFATE ION | | Authors: | Maier, T, Rossman, M, Saenger, W. | | Deposit date: | 2007-09-18 | | Release date: | 2008-04-29 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structures of human saposins C and d: implications for lipid recognition and membrane interactions.
Structure, 16, 2008
|
|
5JIW
 
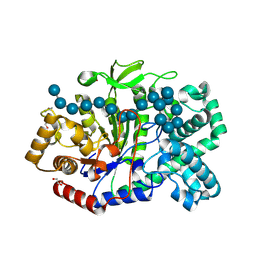 | | Crystal structure of Thermus aquaticus amylomaltase (GH77) in complex with a 34-meric cycloamylose | | Descriptor: | 1,2-ETHANEDIOL, 4-alpha-glucanotransferase, CARBONATE ION, ... | | Authors: | Roth, C, Bexten, N, Weizenmann, N, Saenger, T, Maier, T, Zimmermann, W, Straeter, N. | | Deposit date: | 2016-04-22 | | Release date: | 2017-01-25 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Amylose recognition and ring-size determination of amylomaltase.
Sci Adv, 3, 2017
|
|
4WO7
 
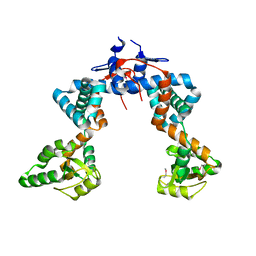 | |
8RQH
 
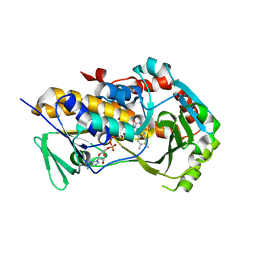 | | Crystal Structure of the flavoprotein monooxygenase TrlE from Streptomyces cyaneofuscatus Soc7 | | Descriptor: | CHLORIDE ION, FLAVIN-ADENINE DINUCLEOTIDE, SULFATE ION, ... | | Authors: | Sowa, S.T, Hoeing, L.S, Jakob, R.P, Maier, T, Teufel, R. | | Deposit date: | 2024-01-18 | | Release date: | 2024-05-15 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Biosynthesis of the bacterial antibiotic 3,7-dihydroxytropolone through enzymatic salvaging of catabolic shunt products.
Chem Sci, 15, 2024
|
|
4CW5
 
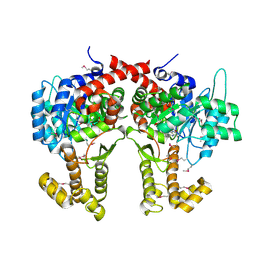 | |
4V5B
 
 | | Structure of PDF binding helix in complex with the ribosome. | | Descriptor: | 16S RIBOSOMAL RNA, 23S RIBOSOMAL RNA, 30S RIBOSOMAL PROTEIN S10, ... | | Authors: | Bingel-Erlenmeyer, R, Kohler, R, Kramer, G, Sandikci, A, Antolic, S, Maier, T, Schaffitzel, C, Wiedmann, B, Bukau, B, Ban, N. | | Deposit date: | 2007-11-22 | | Release date: | 2014-07-09 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (3.74 Å) | | Cite: | A Peptide Deformylase-Ribosome Complex Reveals Mechanism of Nascent Chain Processing.
Nature, 452, 2008
|
|
8CPQ
 
 | |
6YL3
 
 | | High resolution cryo-EM structure of urease from the pathogen Yersinia enterocolitica | | Descriptor: | NICKEL (II) ION, Urease subunit alpha, Urease subunit beta, ... | | Authors: | Righetto, R.D, Anton, L, Adaixo, R, Jakob, R, Zivanov, J, Mahi, M.A, Ringler, P, Schwede, T, Maier, T, Stahlberg, H. | | Deposit date: | 2020-04-06 | | Release date: | 2020-05-06 | | Last modified: | 2025-04-09 | | Method: | ELECTRON MICROSCOPY (1.98 Å) | | Cite: | High-resolution cryo-EM structure of urease from the pathogen Yersinia enterocolitica.
Nat Commun, 11, 2020
|
|
6EYK
 
 | | E-selectin lectin, EGF-like and two SCR domains complexed with glycomimetic ligand NV355 | | Descriptor: | (2~{S})-3-cyclohexyl-2-[(2~{R},3~{S},4~{S},5~{R},6~{R})-2-(hydroxymethyl)-6-[(1~{R},2~{R},3~{S})-3-methyl-2-[(2~{R},3~{S},4~{R},5~{S},6~{R})-3,4,5-tris(oxidanyl)-6-(trifluoromethyl)oxan-2-yl]oxy-cyclohexyl]oxy-3,5-bis(oxidanyl)oxan-4-yl]oxy-propanoic acid, 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Jakob, R.P, Zihlmann, P, Preston, R.C, Varga, N, Ernst, B, Maier, T. | | Deposit date: | 2017-11-13 | | Release date: | 2018-11-21 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | Strengthening an Intramolecular Non-Classical Hydrogen Bond to Get in Shape for Binding.
Angew.Chem.Int.Ed.Engl., 63, 2024
|
|
6EYJ
 
 | | E-selectin lectin, EGF-like and two SCR domains complexed with glycomimetic ligand NV354 | | Descriptor: | (2~{S})-3-cyclohexyl-2-[(2~{R},3~{S},4~{S},5~{R},6~{R})-2-(hydroxymethyl)-3,5-bis(oxidanyl)-6-[(1~{R},2~{R})-2-[(2~{R},3~{S},4~{R},5~{S},6~{R})-3,4,5-tris(oxidanyl)-6-(trifluoromethyl)oxan-2-yl]oxycyclohexyl]oxy-oxan-4-yl]oxy-propanoic acid, 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Jakob, R.P, Zihlmann, P, Preston, R.C, Varga, N, Ernst, B, Maier, T. | | Deposit date: | 2017-11-13 | | Release date: | 2018-11-21 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Strengthening an Intramolecular Non-Classical Hydrogen Bond to Get in Shape for Binding.
Angew.Chem.Int.Ed.Engl., 63, 2024
|
|
8B7S
 
 | | Crystal structure of the Chloramphenicol-inactivating oxidoreductase from Novosphingobium sp | | Descriptor: | Chloramphenicol-inactivating oxidoreductase, FLAVIN-ADENINE DINUCLEOTIDE | | Authors: | Zhang, L, Toplak, M, Saleem-Batcha, R, Hoeing, L, Jakob, R.P, Jehmlich, N, von Bergen, M, Maier, T, Teufel, R. | | Deposit date: | 2022-10-03 | | Release date: | 2022-11-16 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Bacterial Dehydrogenases Facilitate Oxidative Inactivation and Bioremediation of Chloramphenicol.
Chembiochem, 24, 2023
|
|
3J9G
 
 | | Atomic model of the VipA/VipB, the type six secretion system contractile sheath of Vibrio cholerae from cryo-EM | | Descriptor: | VipA, VipB | | Authors: | Kudryashev, M, Wang, R.Y.-R, Brackmann, M, Scherer, S, Maier, T, Baker, D, DiMaio, F, Stahlberg, H, Egelman, E.H, Basler, M. | | Deposit date: | 2015-01-16 | | Release date: | 2015-03-11 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structure of the Type VI Secretion System Contractile Sheath.
Cell(Cambridge,Mass.), 160, 2015
|
|
8R5M
 
 | | E-selectin complexed with glycomimetic ligand DS0567 | | Descriptor: | (2~{S})-3-cyclohexyl-2-[(2~{R},3~{R},4~{S},5~{S},6~{R})-2-[(1~{R},2~{R},3~{S})-3-ethyl-2-[(2~{S},3~{S},4~{R},5~{S},6~{S})-6-methyl-3,4,5-tris(oxidanyl)oxan-2-yl]oxy-cyclohexyl]oxy-6-(hydroxymethyl)-5-oxidanyl-3-(phenylcarbonyloxy)oxan-4-yl]oxy-propanoic acid, 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Jakob, R.P, Ernst, B, Maier, T. | | Deposit date: | 2023-11-17 | | Release date: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | Analogues of the pan-selectin antagonist rivipansel (GMI-1070).
Eur.J.Med.Chem., 272, 2024
|
|
8R5L
 
 | | E-selectin complexed with glycomimetic ligand BW850 | | Descriptor: | (2~{S})-3-cyclohexyl-2-[(2~{R},3~{R},4~{S},5~{S},6~{R})-2-[(1~{R},2~{R},3~{S},5~{R})-3-ethyl-2-[(2~{S},3~{S},4~{R},5~{S},6~{S})-6-methyl-3,4,5-tris(oxidanyl)oxan-2-yl]oxy-5-[3-[1-[3-oxidanylidene-3-[(3,6,8-trisulfonaphthalen-1-yl)amino]propyl]-1,2,3-triazol-4-yl]propylcarbamoyl]cyclohexyl]oxy-6-(hydroxymethyl)-5-oxidanyl-3-(phenylcarbonyloxy)oxan-4-yl]oxy-propanoic acid, 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Jakob, R.P, Ernst, B, Maier, T. | | Deposit date: | 2023-11-17 | | Release date: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Analogues of the pan-selectin antagonist rivipansel (GMI-1070).
Eur.J.Med.Chem., 272, 2024
|
|
6EYI
 
 | | E-selectin lectin, EGF-like and two SCR domains complexed with glycomimetic ligand BW69669 | | Descriptor: | (2~{R})-3-cyclohexyl-2-[(2~{R},3~{S},4~{S},5~{R},6~{R})-2-(hydroxymethyl)-6-[(1~{R},2~{R})-2-[(2~{S},3~{S},4~{R},5~{S},6~{S})-6-methyl-3,4,5-tris(oxidanyl)oxan-2-yl]oxycyclohexyl]oxy-3,5-bis(oxidanyl)oxan-4-yl]oxy-propanoic acid, 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Jakob, R.P, Zihlmann, P, Preston, R.C, Varga, N, Ernst, B, Maier, T. | | Deposit date: | 2017-11-13 | | Release date: | 2018-11-21 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | E-selectin lectin with different glycomimetic ligands
To Be Published
|
|
6FIJ
 
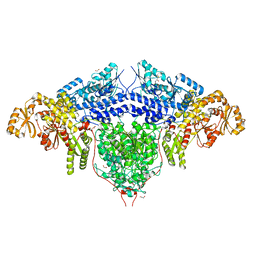 | | Structure of the loading/condensing region (SAT-KS-MAT) of the cercosporin fungal non-reducing polyketide synthase (NR-PKS) CTB1 | | Descriptor: | 1,2-ETHANEDIOL, 2,3-DIHYDROXY-1,4-DITHIOBUTANE, GLYCEROL, ... | | Authors: | Herbst, D.A, Jakob, R.P, Townsend, C.A, Maier, T. | | Deposit date: | 2018-01-18 | | Release date: | 2018-03-21 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.77 Å) | | Cite: | The structural organization of substrate loading in iterative polyketide synthases.
Nat. Chem. Biol., 14, 2018
|
|
6QGX
 
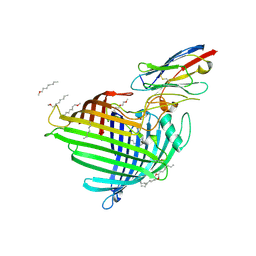 | | Crystal structure of E.coli BamA beta-barrel in complex with nanobody F7 | | Descriptor: | (HYDROXYETHYLOXY)TRI(ETHYLOXY)OCTANE, NanoF7, Outer membrane protein assembly factor BamA | | Authors: | Hartmann, J.-B, Kaur, H, Jakob, R.P, Zahn, M, Zimmermann, I, Seeger, M, Maier, T, Hiller, S. | | Deposit date: | 2019-01-14 | | Release date: | 2019-06-26 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Identification of conformation-selective nanobodies against the membrane protein insertase BamA by an integrated structural biology approach.
J.Biomol.Nmr, 73, 2019
|
|
