5IBO
 
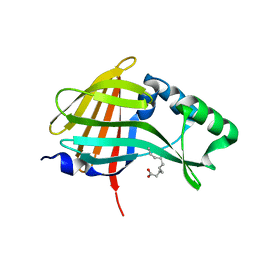 | | 1.95A resolution structure of NanoLuc luciferase | | Descriptor: | DECANOIC ACID, Oplophorus-luciferin 2-monooxygenase catalytic subunit | | Authors: | Lovell, S, Mehzabeen, N, Battaile, K.P, Wood, M.G, Encell, L.P, Wood, K.V. | | Deposit date: | 2016-02-22 | | Release date: | 2016-04-13 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | To be determined
To be published
|
|
9N1S
 
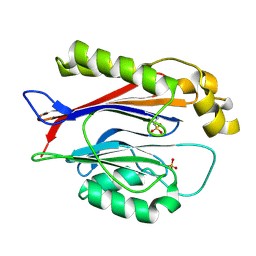 | | Crystal structure of the Transport and Golgi Organization protein 2 Homolog (TANGO2) tetragonal form | | Descriptor: | CHLORIDE ION, PHOSPHATE ION, Transport and Golgi organization protein 2 homolog | | Authors: | Lovell, S, Cooper, A, Powers, A, Battaile, K.P, Moshen, A.W, Ghaloul-Gonzalez, L. | | Deposit date: | 2025-01-27 | | Release date: | 2025-04-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of the Transport and Golgi Organization protein 2 Homolog (TANGO2)
To Be Published
|
|
9BWA
 
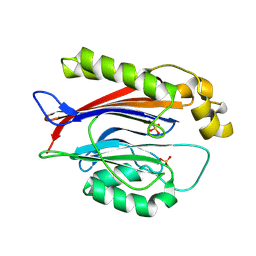 | | Crystal structure of the Transport and Golgi Organization protein 2 Homolog (TANGO2) | | Descriptor: | PHOSPHATE ION, Transport and Golgi organization protein 2 homolog | | Authors: | Lovell, S, Cooper, A, Powers, A, Battaile, K.P, Mohsen, A.-W, Ghaloul-Gonzalez, L. | | Deposit date: | 2024-05-21 | | Release date: | 2025-04-30 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of the Transport and Golgi Organization protein 2 Homolog (TANGO2)
To Be Published
|
|
7TPM
 
 | | Structure of the outer-membrane lipoprotein carrier protein (LolA) from Borrelia burgdorferi | | Descriptor: | 2-{2-[2-(2-{2-[2-(2-ETHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHOXY]-ETHOXY}-ETHANOL, CHLORIDE ION, Outer membrane lipoprotein carrier protein LolA, ... | | Authors: | Lovell, S, Kashipathy, M.M, Battaile, K.P, Zueckert, W.R. | | Deposit date: | 2022-01-25 | | Release date: | 2023-03-15 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure of the outer-membrane lipoprotein carrier protein (LolA) from Borrelia burgdorferi
To be published
|
|
7RT0
 
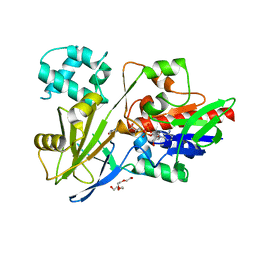 | |
5C98
 
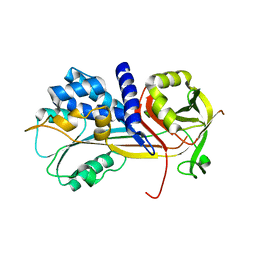 | | 1.45A resolution structure of SRPN18 from Anopheles gambiae | | Descriptor: | AGAP007691-PB | | Authors: | Lovell, S, Battaile, K.P, Gulley, M, Zhang, X, Meekins, D.A, Gao, F.P, Michel, K. | | Deposit date: | 2015-06-26 | | Release date: | 2016-09-14 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | 1.45 angstrom resolution structure of SRPN18 from the malaria vector Anopheles gambiae.
Acta Crystallogr F Struct Biol Commun, 72, 2016
|
|
3CXK
 
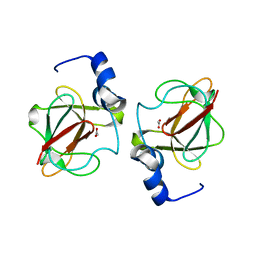 | | 1.7 A Crystal structure of methionine-R-sulfoxide reductase from Burkholderia pseudomallei: crystallization in a microfluidic crystal card. | | Descriptor: | ACETATE ION, Methionine-R-sulfoxide reductase, ZINC ION | | Authors: | Lovell, S, Gerdts, C, Staker, B, Craigen, D, Stewart, L, Accelerated Technologies Center for Gene to 3D Structure (ATCG3D), Seattle Structural Genomics Center for Infectious Disease (SSGCID) | | Deposit date: | 2008-04-24 | | Release date: | 2008-05-27 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The plug-based nanovolume Microcapillary Protein Crystallization System (MPCS).
Acta Crystallogr.,Sect.D, 64, 2008
|
|
6XMK
 
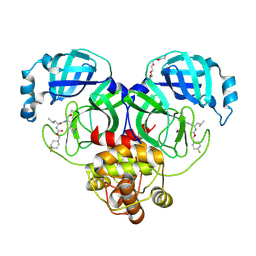 | | 1.70 A resolution structure of SARS-CoV-2 3CL protease in complex with inhibitor 7j | | Descriptor: | (1S,2S)-2-[(N-{[(4,4-difluorocyclohexyl)methoxy]carbonyl}-L-leucyl)amino]-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid, 3C-like proteinase, TETRAETHYLENE GLYCOL | | Authors: | Lovell, S, Kashipathy, M.M, Battaile, K.P, Rathnayake, A.D, Zheng, J, Kim, Y, Nguyen, H.N, Chang, K.O, Groutas, W.C. | | Deposit date: | 2020-06-30 | | Release date: | 2020-07-08 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | 3C-like protease inhibitors block coronavirus replication in vitro and improve survival in MERS-CoV-infected mice.
Sci Transl Med, 12, 2020
|
|
3UR6
 
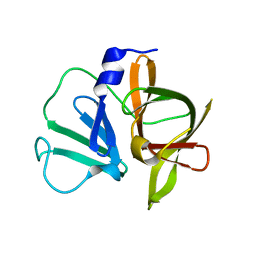 | | 1.5A resolution structure of apo Norwalk Virus Protease | | Descriptor: | 3C-like protease | | Authors: | Lovell, S, Battaile, K.P, Kim, Y, Tiew, K.C, Mandadapu, S.R, Alliston, K.R, Groutas, W.C, Chang, K.O. | | Deposit date: | 2011-11-21 | | Release date: | 2012-09-05 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Broad-Spectrum Antivirals against 3C or 3C-Like Proteases of Picornaviruses, Noroviruses, and Coronaviruses.
J.Virol., 86, 2012
|
|
3UR9
 
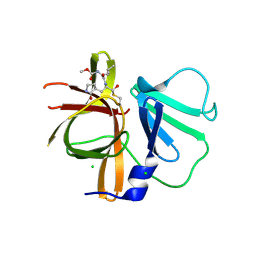 | | 1.65A resolution structure of Norwalk Virus Protease Containing a covalently bound dipeptidyl inhibitor | | Descriptor: | (1S,2S)-2-({N-[(benzyloxy)carbonyl]-L-leucyl}amino)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid, 3C-like protease, CHLORIDE ION | | Authors: | Lovell, S, Battaile, K.P, Kim, Y, Tiew, K.C, Mandadapu, S.R, Alliston, K.R, Groutas, W.C, Chang, K.O. | | Deposit date: | 2011-11-21 | | Release date: | 2012-09-05 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Broad-Spectrum Antivirals against 3C or 3C-Like Proteases of Picornaviruses, Noroviruses, and Coronaviruses.
J.Virol., 86, 2012
|
|
4XBD
 
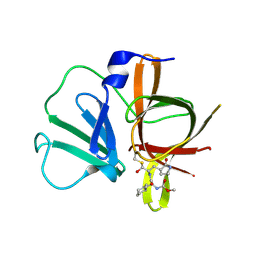 | | 1.45A resolution structure of Norovirus 3CL protease complex with a covalently bound dipeptidyl inhibitor (1R,2S)-2-({N-[(benzyloxy)carbonyl]-3-cyclohexyl-L-alanyl}amino)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid (Orthorhombic P Form) | | Descriptor: | (1R,2S)-2-({N-[(benzyloxy)carbonyl]-3-cyclohexyl-L-alanyl}amino)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid, 3C-LIKE PROTEASE | | Authors: | Lovell, S, Battaile, K.P, Mehzabeen, N, Kankanamalage, A.C.G, Kim, Y, Weerawarna, P.M, Uy, R.A.Z, Damalanka, V.C, Mandadapu, S.R, Alliston, K.R, Groutas, W.C, Chang, K.-O. | | Deposit date: | 2014-12-16 | | Release date: | 2015-03-25 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Structure-Guided Design and Optimization of Dipeptidyl Inhibitors of Norovirus 3CL Protease. Structure-Activity Relationships and Biochemical, X-ray Crystallographic, Cell-Based, and In Vivo Studies.
J.Med.Chem., 58, 2015
|
|
9BK3
 
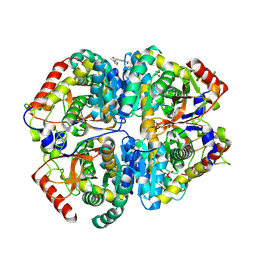 | | Crystal structure of Lactate dehydrogenase in complex with 4-((4-(1-methyl-1H-imidazole-2-carbonyl)phenyl)amino)-4-oxo-2-(4-(trifluoromethyl)phenyl)butanoic acid (R-enantiomer, orthorhombic P form) | | Descriptor: | (2R)-4-[4-(1-methyl-1H-imidazole-2-carbonyl)anilino]-4-oxo-2-[4-(trifluoromethyl)phenyl]butanoic acid, CHLORIDE ION, L-lactate dehydrogenase A chain, ... | | Authors: | Lovell, S, Cooper, A, Battaile, K.P, Sharma, H. | | Deposit date: | 2024-04-26 | | Release date: | 2024-07-10 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Synthesis and biological characterization of an orally bioavailable lactate dehydrogenase-A inhibitor against pancreatic cancer.
Eur.J.Med.Chem., 275, 2024
|
|
9BK2
 
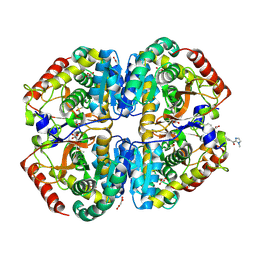 | | Crystal structure of Lactate dehydrogenase in complex with 4-((4-(1-methyl-1H-imidazole-2-carbonyl)phenyl)amino)-4-oxo-2-(4-(trifluoromethyl)phenyl)butanoic acid (S-enantiomer, monoclinic P form) | | Descriptor: | (2S)-4-[4-(1-methyl-1H-imidazole-2-carbonyl)anilino]-4-oxo-2-[4-(trifluoromethyl)phenyl]butanoic acid, DIMETHYL SULFOXIDE, L-lactate dehydrogenase A chain, ... | | Authors: | Lovell, S, Cooper, A, Battaile, K.P, Sharma, H. | | Deposit date: | 2024-04-26 | | Release date: | 2024-07-10 | | Last modified: | 2024-09-04 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Synthesis and biological characterization of an orally bioavailable lactate dehydrogenase-A inhibitor against pancreatic cancer.
Eur.J.Med.Chem., 275, 2024
|
|
4XBC
 
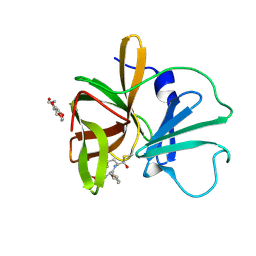 | | 1.60 A resolution structure of Norovirus 3CL protease complex with a covalently bound dipeptidyl inhibitor (1R,2S)-2-({N-[(benzyloxy)carbonyl]-3-cyclohexyl-L-alanyl}amino)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid (Hexagonal Form) | | Descriptor: | (1R,2S)-2-({N-[(benzyloxy)carbonyl]-3-cyclohexyl-L-alanyl}amino)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propane-1-sulfonic acid, 3C-LIKE PROTEASE, TETRAETHYLENE GLYCOL | | Authors: | Lovell, S, Battaile, K.P, Mehzabeen, N, Kankanamalage, A.C.G, Kim, Y, Weerawarna, P.M, Uy, R.A.Z, Damalanka, V.C, Mandadapu, S.R, Alliston, K.R, Groutas, W.C, Chang, K.-O. | | Deposit date: | 2014-12-16 | | Release date: | 2015-03-25 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure-Guided Design and Optimization of Dipeptidyl Inhibitors of Norovirus 3CL Protease. Structure-Activity Relationships and Biochemical, X-ray Crystallographic, Cell-Based, and In Vivo Studies.
J.Med.Chem., 58, 2015
|
|
4XBB
 
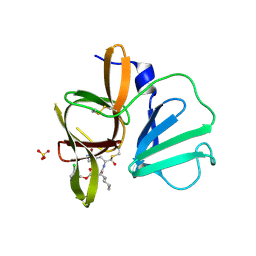 | | 1.85A resolution structure of Norovirus 3CL protease complex with a covalently bound dipeptidyl inhibitor diethyl [(1R,2S)-2-[(N-{[(3-chlorobenzyl)oxy]carbonyl}-3-cyclohexyl-L-alanyl)amino]-1-hydroxy-3-(2-oxo-2H-pyrrol-3-yl)propyl]phosphonate | | Descriptor: | 3C-LIKE PROTEASE, SULFATE ION, diethyl [(1R,2S)-2-[(N-{[(3-chlorobenzyl)oxy]carbonyl}-3-cyclohexyl-L-alanyl)amino]-1-hydroxy-3-(2-oxo-2H-pyrrol-3-yl)propyl]phosphonate | | Authors: | Lovell, S, Battaile, K.P, Mehzabeen, N, Kankanamalage, A.C.G, Kim, Y, Weerawarna, P.M, Uy, R.A.Z, Damalanka, V.C, Mandadapu, S.R, Alliston, K.R, Groutas, W.C, Chang, K.-O. | | Deposit date: | 2014-12-16 | | Release date: | 2015-03-25 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structure-Guided Design and Optimization of Dipeptidyl Inhibitors of Norovirus 3CL Protease. Structure-Activity Relationships and Biochemical, X-ray Crystallographic, Cell-Based, and In Vivo Studies.
J.Med.Chem., 58, 2015
|
|
6WOJ
 
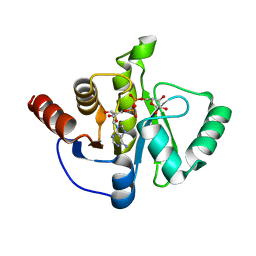 | | Structure of the SARS-CoV-2 macrodomain (NSP3) in complex with ADP-ribose | | Descriptor: | ADENOSINE-5-DIPHOSPHORIBOSE, Non-structural protein 3 | | Authors: | Lovell, S, Kashipathy, M.M, Battaile, K.P, Gao, F.P, Fehr, A.R. | | Deposit date: | 2020-04-24 | | Release date: | 2020-05-06 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The SARS-CoV-2 Conserved Macrodomain Is a Mono-ADP-Ribosylhydrolase.
J.Virol., 95, 2021
|
|
5KGN
 
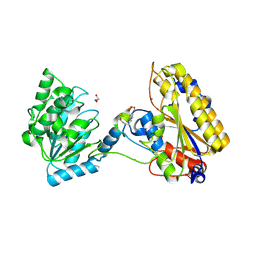 | | 1.95A resolution structure of independent phosphoglycerate mutase from C. elegans in complex with a macrocyclic peptide inhibitor (2d) | | Descriptor: | 2,3-bisphosphoglycerate-independent phosphoglycerate mutase, CHLORIDE ION, GLYCEROL, ... | | Authors: | Lovell, S, Mehzabeen, N, Battaile, K.P, Yu, H, Dranchak, P, MacArthur, R, Li, Z, Carlow, T, Suga, H, Inglese, J. | | Deposit date: | 2016-06-13 | | Release date: | 2017-04-05 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Macrocycle peptides delineate locked-open inhibition mechanism for microorganism phosphoglycerate mutases.
Nat Commun, 8, 2017
|
|
5KGM
 
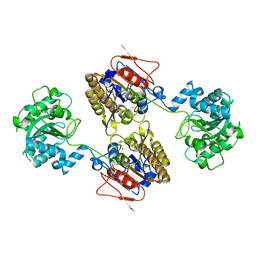 | | 2.95A resolution structure of Apo independent phosphoglycerate mutase from C. elegans (monoclinic form) | | Descriptor: | 2,3-bisphosphoglycerate-independent phosphoglycerate mutase, CHLORIDE ION, MANGANESE (II) ION, ... | | Authors: | Lovell, S, Mehzabeen, N, Battaile, K.P, Yu, H, Dranchak, P, MacArthur, R, Li, Z, Carlow, T, Suga, H, Inglese, J. | | Deposit date: | 2016-06-13 | | Release date: | 2017-04-05 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Macrocycle peptides delineate locked-open inhibition mechanism for microorganism phosphoglycerate mutases.
Nat Commun, 8, 2017
|
|
5KGL
 
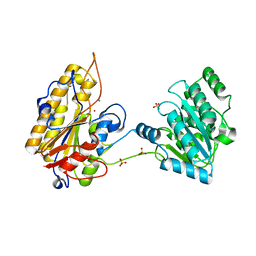 | | 2.45A resolution structure of Apo independent phosphoglycerate mutase from C. elegans (orthorhombic form) | | Descriptor: | 2,3-bisphosphoglycerate-independent phosphoglycerate mutase, CHLORIDE ION, MANGANESE (II) ION, ... | | Authors: | Lovell, S, Mehzabeen, N, Battaile, K.P, Yu, H, Dranchak, P, MacArthur, R, Li, Z, Carlow, T, Suga, H, Inglese, J. | | Deposit date: | 2016-06-13 | | Release date: | 2017-04-05 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Macrocycle peptides delineate locked-open inhibition mechanism for microorganism phosphoglycerate mutases.
Nat Commun, 8, 2017
|
|
9BTS
 
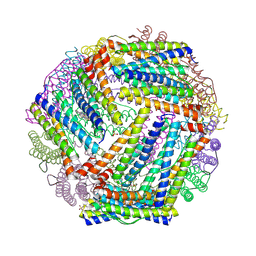 | | Crystal structure of the bacterioferritin (Bfr) and ferritin (Ftn) heterooligomer complex from Acinetobacter baumannii | | Descriptor: | Bacterioferritin (Bfr), Ferritin (Ftn), PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Lovell, S, Liu, L, Battaile, K.P, Rivera, M. | | Deposit date: | 2024-05-15 | | Release date: | 2024-08-14 | | Last modified: | 2024-08-21 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | The crystal structure of Acinetobacter baumannii bacterioferritin reveals a heteropolymer of bacterioferritin and ferritin subunits.
Sci Rep, 14, 2024
|
|
5D8Q
 
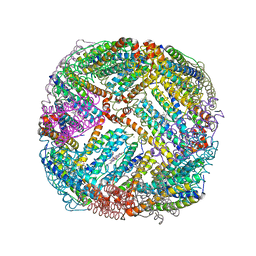 | | 2.20A resolution structure of BfrB (L68A) from Pseudomonas aeruginosa | | Descriptor: | ARSENIC, Ferroxidase, MAGNESIUM ION, ... | | Authors: | Lovell, S, Battaile, K.P, Wang, Y, Yao, H, Rivera, M. | | Deposit date: | 2015-08-17 | | Release date: | 2015-09-23 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Characterization of the Bacterioferritin/Bacterioferritin Associated Ferredoxin Protein-Protein Interaction in Solution and Determination of Binding Energy Hot Spots.
Biochemistry, 54, 2015
|
|
5D8O
 
 | | 1.90A resolution structure of BfrB (wild-type, C2221 form) from Pseudomonas aeruginosa | | Descriptor: | Ferroxidase, POTASSIUM ION, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Lovell, S, Battaile, K.P, Wang, Y, Yao, H, Rivera, M. | | Deposit date: | 2015-08-17 | | Release date: | 2015-09-23 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Characterization of the Bacterioferritin/Bacterioferritin Associated Ferredoxin Protein-Protein Interaction in Solution and Determination of Binding Energy Hot Spots.
Biochemistry, 54, 2015
|
|
5DGJ
 
 | | 1.0A resolution structure of Norovirus 3CL protease in complex an oxadiazole-based, cell permeable macrocyclic (20-mer) inhibitor | | Descriptor: | 3C-LIKE PROTEASE, tert-butyl [(4S,7S,10S)-7-(cyclohexylmethyl)-10-(hydroxymethyl)-5,8,13-trioxo-22-oxa-6,9,14,20,21-pentaazabicyclo[17.2.1]docosa-1(21),19-dien-4-yl]carbamate | | Authors: | Lovell, S, Battaile, K.P, Mehzabeen, N, Damalanka, V.C, Kim, Y, Alliston, K.R, Weerawarna, P.M, Kankanamalage, A.C.G, Lushington, G.H, Chang, K.-O, Groutas, W.C. | | Deposit date: | 2015-08-27 | | Release date: | 2016-02-10 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Oxadiazole-Based Cell Permeable Macrocyclic Transition State Inhibitors of Norovirus 3CL Protease.
J.Med.Chem., 59, 2016
|
|
5D8Y
 
 | | 2.05A resolution structure of iron bound BfrB (L68A E81A) from Pseudomonas aeruginosa | | Descriptor: | ACETATE ION, FE (II) ION, Ferroxidase, ... | | Authors: | Lovell, S, Battaile, K.P, Wang, Y, Yao, H, Rivera, M. | | Deposit date: | 2015-08-18 | | Release date: | 2015-09-23 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Characterization of the Bacterioferritin/Bacterioferritin Associated Ferredoxin Protein-Protein Interaction in Solution and Determination of Binding Energy Hot Spots.
Biochemistry, 54, 2015
|
|
5D8P
 
 | | 2.35A resolution structure of iron bound BfrB (wild-type, C2221 form) from Pseudomonas aeruginosa | | Descriptor: | ACETATE ION, FE (II) ION, Ferroxidase, ... | | Authors: | Lovell, S, Battaile, K.P, Wang, Y, Yao, H, Rivera, M. | | Deposit date: | 2015-08-17 | | Release date: | 2015-09-23 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Characterization of the Bacterioferritin/Bacterioferritin Associated Ferredoxin Protein-Protein Interaction in Solution and Determination of Binding Energy Hot Spots.
Biochemistry, 54, 2015
|
|
