3KZA
 
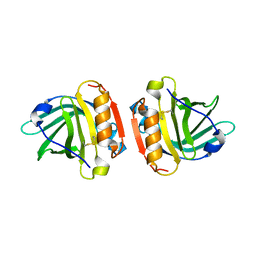 | | Crystal structure of Gyuba, a patched chimera of b-lactglobulin | | Descriptor: | Beta-lactoglobulin | | Authors: | Tsuge, H, Ohtomo, H, Utsunomiya, H, Konuma, T, Ikeguchi, M. | | Deposit date: | 2009-12-08 | | Release date: | 2010-12-22 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure and stability of Gyuba, a patched chimera of b-lactoglobulin
Protein Sci., 20, 2011
|
|
2NOO
 
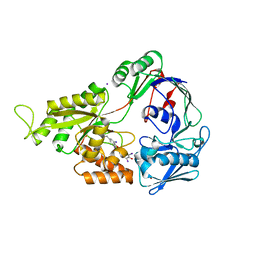 | | Crystal Structure of Mutant NikA | | Descriptor: | IODIDE ION, NICKEL (II) ION, Nickel-binding periplasmic protein | | Authors: | Addy, C, Ohara, M, Kawai, F, Kidera, A, Ikeguchi, M, Fuchigami, S, Osawa, M, Shimada, I, Park, S.Y, Tame, J.R.H, Heddle, J.G. | | Deposit date: | 2006-10-26 | | Release date: | 2007-01-23 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Nickel binding to NikA: an additional binding site reconciles spectroscopy, calorimetry and crystallography.
Acta Crystallogr.,Sect.D, 63, 2007
|
|
1K85
 
 | | Solution structure of the fibronectin type III domain from Bacillus circulans WL-12 Chitinase A1. | | Descriptor: | CHITINASE A1 | | Authors: | Jee, J.G, Ikegami, T, Hashimoto, M, Kawabata, T, Ikeguchi, M, Watanabe, T, Shirakawa, M. | | Deposit date: | 2001-10-23 | | Release date: | 2002-12-18 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of the Fibronectin Type III Domain
from Bacillus circulans WL-12 Chitinase A1
J.Biol.Chem., 277, 2002
|
|
2Z33
 
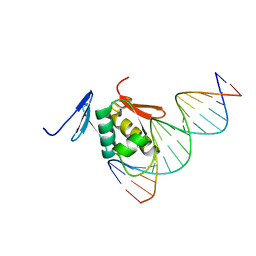 | | Solution structure of the DNA complex of PhoB DNA-binding/transactivation Domain | | Descriptor: | 5'-D(*AP*CP*AP*GP*AP*TP*TP*TP*AP*TP*GP*AP*CP*AP*GP*T)-3', 5'-D(*AP*CP*TP*GP*TP*CP*AP*TP*AP*AP*AP*TP*CP*TP*GP*T)-3', Phosphate regulon transcriptional regulatory protein phoB | | Authors: | Yamane, T, Okamura, H, Ikeguchi, M, Nishimura, Y, Kidera, A. | | Deposit date: | 2007-05-31 | | Release date: | 2008-04-22 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Water-mediated interactions between DNA and PhoB DNA-binding/transactivation domain: NMR-restrained molecular dynamics in explicit water environment.
Proteins, 71, 2008
|
|
3A57
 
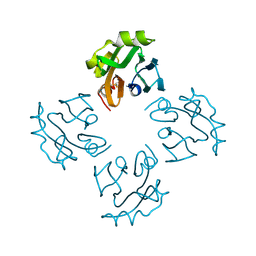 | | Crystal structure of Thermostable Direct Hemolysin | | Descriptor: | Thermostable direct hemolysin 2 | | Authors: | Hashimoto, H, Yanagihara, I, Nakahira, K, Hamada, D, Ikegami, T, Mayanagi, K, Kaieda, S, Fukui, T, Ohnishi, K, Kajiyama, S, Yamane, T, Ikeguchi, M, Honda, T, Shimizu, T, Sato, M. | | Deposit date: | 2009-08-03 | | Release date: | 2010-03-31 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure and functional characterization of Vibrio parahaemolyticus thermostable direct hemolysin
J.Biol.Chem., 285, 2010
|
|
3VIQ
 
 | | Crystal structure of Swi5-Sfr1 complex from fission yeast | | Descriptor: | GLYCEROL, Mating-type switching protein swi5, NITRATE ION, ... | | Authors: | Kuwabara, N, Murayama, Y, Hashimoto, H, Kokabu, Y, Ikeguchi, M, Sato, M, Mayanagi, K, Tsutsui, Y, Iwasaki, H, Shimizu, T. | | Deposit date: | 2011-10-06 | | Release date: | 2012-08-22 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Mechanistic insights into the activation of Rad51-mediated strand exchange from the structure of a recombination activator, the Swi5-Sfr1 complex
Structure, 20, 2012
|
|
2RPQ
 
 | | Solution Structure of a SUMO-interacting motif of MBD1-containing chromatin-associated factor 1 bound to SUMO-3 | | Descriptor: | Activating transcription factor 7-interacting protein 1, Small ubiquitin-related modifier 2 | | Authors: | Sekiyama, N, Ikegami, T, Yamane, T, Ikeguchi, M, Uchimura, Y, Baba, D, Ariyoshi, M, Tochio, H, Saitoh, H, Shirakawa, M. | | Deposit date: | 2008-07-07 | | Release date: | 2008-10-07 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structure of the small ubiquitin-like modifier (SUMO)-interacting motif of MBD1-containing chromatin-associated factor 1 bound to SUMO-3
J.Biol.Chem., 283, 2008
|
|
2RVQ
 
 | | Solution structure of the isolated histone H2A-H2B heterodimer | | Descriptor: | Histone H2A type 1-B/E, Histone H2B type 1-J | | Authors: | Moriwaki, Y, Yamane, T, Ohtomo, H, Ikeguchi, M, Kurita, J, Sato, M, Nagadoi, A, Shimojo, H, Nishimura, Y. | | Deposit date: | 2016-03-28 | | Release date: | 2016-05-25 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the isolated histone H2A-H2B heterodimer
Sci Rep, 6, 2016
|
|
2RQ7
 
 | | Solution structure of the epsilon subunit chimera combining the N-terminal beta-sandwich domain from T. Elongatus bp-1 f1 and the C-terminal alpha-helical domain from spinach chloroplast F1 | | Descriptor: | ATP synthase epsilon chain,ATP synthase epsilon chain, chloroplastic | | Authors: | Yagi, H, Konno, H, Murakami-Fuse, T, Oroguchi, H, Akutsu, T, Ikeguchi, M, Hisabori, T. | | Deposit date: | 2009-03-03 | | Release date: | 2010-01-12 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structural and functional analysis of the intrinsic inhibitor subunit epsilon of F1-ATPase from photosynthetic organisms.
Biochem.J., 425, 2010
|
|
2RQ6
 
 | | Solution structure of the epsilon subunit of the F1-atpase from thermosynechococcus elongatus BP-1 | | Descriptor: | ATP synthase epsilon chain | | Authors: | Yagi, H, Konno, H, Murakami-Fuse, T, Oroguchi, H, Akutsu, T, Ikeguchi, M, Hisabori, T. | | Deposit date: | 2009-03-03 | | Release date: | 2010-01-12 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Structural and functional analysis of the intrinsic inhibitor subunit epsilon of F1-ATPase from photosynthetic organisms.
Biochem.J., 425, 2010
|
|
6LVY
 
 | | Crystal structure of TLR7/Cpd-2 (SM-360320) complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 6-azanyl-2-(2-methoxyethoxy)-9-(phenylmethyl)-7H-purin-8-one, ... | | Authors: | Zhang, Z, Ohto, U, Shimizu, T. | | Deposit date: | 2020-02-06 | | Release date: | 2020-11-11 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural analysis reveals TLR7 dynamics underlying antagonism.
Nat Commun, 11, 2020
|
|
7FB7
 
 | | Crystal structure of human UHRF1 TTD in complex with 5-amino-2,4-dimethylpyridine | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 5-amino-2,4-dimethylpyridine, DIMETHYL SULFOXIDE, ... | | Authors: | Kori, S, Arita, K, Yoshimi, S. | | Deposit date: | 2021-07-08 | | Release date: | 2022-01-05 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Structure-based screening combined with computational and biochemical analyses identified the inhibitor targeting the binding of DNA Ligase 1 to UHRF1.
Bioorg.Med.Chem., 52, 2021
|
|
8I7W
 
 | | Cryo-EM structure of GSK256073 bound human hydroxy-carboxylic acid receptor 2 in complex with Gi heterotrimer | | Descriptor: | 8-chloranyl-3-pentyl-7H-purine-2,6-dione, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Park, J.H, Ishimoto, N, Park, S.Y. | | Deposit date: | 2023-02-02 | | Release date: | 2024-02-07 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3.39 Å) | | Cite: | Structural basis for ligand recognition and signaling of hydroxy-carboxylic acid receptor 2.
Nat Commun, 14, 2023
|
|
8I7V
 
 | | Cryo-EM structure of Acipimox bound human hydroxy-carboxylic acid receptor 2 in complex with Gi heterotrimer | | Descriptor: | 5-methyl-4-oxidanyl-pyrazin-4-ium-2-carboxylic acid, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Park, J.H, Ishimoto, N, Park, S.Y. | | Deposit date: | 2023-02-02 | | Release date: | 2024-02-07 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (2.77 Å) | | Cite: | Structural basis for ligand recognition and signaling of hydroxy-carboxylic acid receptor 2.
Nat Commun, 14, 2023
|
|
5WVO
 
 | | Crystal structure of DNMT1 RFTS domain in complex with K18/K23 mono-ubiquitylated histone H3 | | Descriptor: | DNA (cytosine-5)-methyltransferase 1, Histone H3.1, Ubiquitin, ... | | Authors: | Ishiyama, S, Nishiyama, A, Nakanishi, M, Arita, K. | | Deposit date: | 2016-12-28 | | Release date: | 2017-11-15 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.997 Å) | | Cite: | Structure of the Dnmt1 Reader Module Complexed with a Unique Two-Mono-Ubiquitin Mark on Histone H3 Reveals the Basis for DNA Methylation Maintenance
Mol. Cell, 68, 2017
|
|
8WCI
 
 | | Cryo-EM structure of the inhibitor-bound Vo complex from Enterococcus hirae | | Descriptor: | CARDIOLIPIN, N,N-dimethyl-4-(5-methyl-1H-benzimidazol-2-yl)aniline, SODIUM ION, ... | | Authors: | Suzuki, K, Mikuriya, S, Adachi, N, Kawasaki, M, Senda, T, Moriya, T, Murata, T. | | Deposit date: | 2023-09-12 | | Release date: | 2024-10-09 | | Last modified: | 2025-04-23 | | Method: | ELECTRON MICROSCOPY (2.2 Å) | | Cite: | Na + -V-ATPase inhibitor curbs VRE growth and unveils Na + pathway structure.
Nat.Struct.Mol.Biol., 32, 2025
|
|
8H2G
 
 | | Cryo-EM structure of niacin bound human hydroxy-carboxylic acid receptor 2 in complex with Gi heterotrimer | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(i) subunit alpha-1, ... | | Authors: | Park, J.H, Ishimoto, N, Park, S.Y. | | Deposit date: | 2022-10-06 | | Release date: | 2023-10-11 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.01 Å) | | Cite: | Structural basis for ligand recognition and signaling of hydroxy-carboxylic acid receptor 2
Nat Commun, 14, 2023
|
|
1V8Z
 
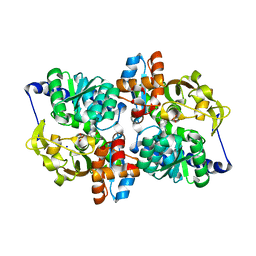 | | X-ray crystal structure of the Tryptophan Synthase b2 Subunit from Hyperthermophile, Pyrococcus furiosus | | Descriptor: | PYRIDOXAL-5'-PHOSPHATE, SODIUM ION, Tryptophan synthase beta chain 1 | | Authors: | Hioki, Y, Ogasahara, K, Lee, S.J, Ma, J, Ishida, M, Yamagata, Y, Matsuura, Y, Ota, M, Kuramitsu, S, Yutani, K, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2004-01-15 | | Release date: | 2005-02-22 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | The crystal structure of the tryptophan synthase beta subunit from the hyperthermophile Pyrococcus furiosus. Investigation of stabilization factors
Eur.J.Biochem., 271, 2004
|
|
6LW0
 
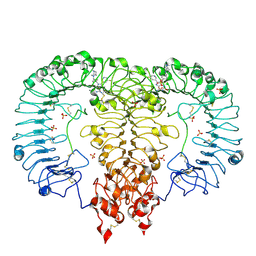 | | Crystal structure of TLR7/Cpd-6 (DSR-139293) complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-ethoxy-8-(5-fluoranylpyridin-3-yl)-9-[[4-[[(1S,4S)-5-methyl-2,5-diazabicyclo[2.2.1]heptan-2-yl]methyl]phenyl]methyl]purin-6-amine, ... | | Authors: | Zhang, Z, Ohto, U, Shimizu, T. | | Deposit date: | 2020-02-06 | | Release date: | 2020-11-11 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural analysis reveals TLR7 dynamics underlying antagonism.
Nat Commun, 11, 2020
|
|
6LW1
 
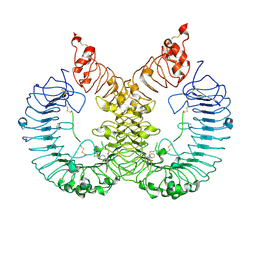 | | Cryo-EM structure of TLR7/Cpd-7 (DSR-139970) complex in open form | | Descriptor: | 2-ethoxy-8-(5-fluoranylpyridin-3-yl)-6-methyl-9-[[4-[[(1S,4S)-5-methyl-2,5-diazabicyclo[2.2.1]heptan-2-yl]methyl]phenyl]methyl]purine, Toll-like receptor 7 | | Authors: | Zhang, Z, Ohto, U, Shimizu, T. | | Deposit date: | 2020-02-06 | | Release date: | 2020-11-11 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Structural analysis reveals TLR7 dynamics underlying antagonism.
Nat Commun, 11, 2020
|
|
6LVX
 
 | | Crystal structure of TLR7/Cpd-1 (SM-374527) complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 6-azanyl-2-butoxy-9-(phenylmethyl)-7H-purin-8-one, ... | | Authors: | Zhang, Z, Ohto, U, Shimizu, T. | | Deposit date: | 2020-02-06 | | Release date: | 2020-11-11 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.77 Å) | | Cite: | Structural analysis reveals TLR7 dynamics underlying antagonism.
Nat Commun, 11, 2020
|
|
6LVZ
 
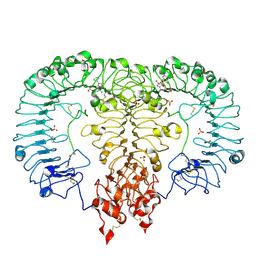 | | Crystal structure of TLR7/Cpd-3 (SM-394830) complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 6-azanyl-2-(2-methoxyethoxy)-9-(pyridin-3-ylmethyl)-7H-purin-8-one, ... | | Authors: | Zhang, Z, Ohto, U, Shimizu, T. | | Deposit date: | 2020-02-06 | | Release date: | 2020-11-11 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.83 Å) | | Cite: | Structural analysis reveals TLR7 dynamics underlying antagonism.
Nat Commun, 11, 2020
|
|
5B41
 
 | | Crystal structure of VDR-LBD complexed with 2-methylidene-19-nor-1a,25-dihydroxyvitamin D3 | | Descriptor: | (1R,3R)-5-(2-((1R,3aS,7aR,E)-1-((R)-6-hydroxy-6-methylheptan-2-yl)-7a-methyloctahydro-4H-inden-4-ylidene)ethylidene)-2- methylenecyclohexane-1,3-diol, Mediator of RNA polymerase II transcription subunit 1, Vitamin D3 receptor | | Authors: | Anami, Y, Itoh, T, Yamamoto, K. | | Deposit date: | 2016-03-23 | | Release date: | 2016-12-07 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Apo- and Antagonist-Binding Structures of Vitamin D Receptor Ligand-Binding Domain Revealed by Hybrid Approach Combining Small-Angle X-ray Scattering and Molecular Dynamics
J.Med.Chem., 59, 2016
|
|
7Y4P
 
 | | Human Plexin A1, extracellular domains 1-4 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Plexin-A1 | | Authors: | Tanaka, T, Neyazaki, M, Nogi, T. | | Deposit date: | 2022-06-15 | | Release date: | 2022-10-19 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Hybrid in vitro/in silico analysis of low-affinity protein-protein interactions that regulate signal transduction by Sema6D.
Protein Sci., 31, 2022
|
|
7Y4O
 
 | | Rat Semaphorin 6D extracellular region | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Semaphorin 6D | | Authors: | Tanaka, T, Neyazaki, M, Nogi, T. | | Deposit date: | 2022-06-15 | | Release date: | 2022-10-19 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Hybrid in vitro/in silico analysis of low-affinity protein-protein interactions that regulate signal transduction by Sema6D.
Protein Sci., 31, 2022
|
|
