1ZS8
 
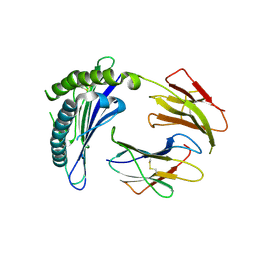 | | Crystal Structure of the Murine MHC Class Ib Molecule M10.5 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Beta-2-microglobulin, histocompatibility 2, ... | | Authors: | Olson, R, Huey-Tubman, K.E, Dulac, C, Bjorkman, P.J. | | Deposit date: | 2005-07-22 | | Release date: | 2005-07-26 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structure of a pheromone receptor-associated MHC molecule with an open and empty groove.
Plos Biol., 3, 2005
|
|
2GSG
 
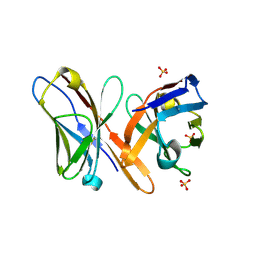 | | Crystal structure of the Fv fragment of a monoclonal antibody specific for poly-glutamine | | Descriptor: | SULFATE ION, monoclonal antibody heavy chain, monoclonal antibody light chain | | Authors: | Li, P, Huey-Tubman, K.E, West Jr, A.P, Bennett, M.J, Bjorkman, P.J. | | Deposit date: | 2006-04-26 | | Release date: | 2007-04-24 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The structure of a polyQ-anti-polyQ complex reveals binding according to a linear lattice model.
Nat.Struct.Mol.Biol., 14, 2007
|
|
7N3E
 
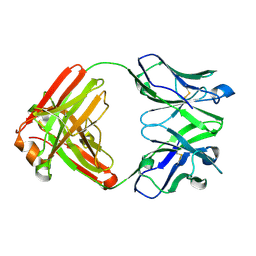 | | Crystal structure of an anti-SARS-CoV-2 human neutralizing antibody Fab fragment C032 | | Descriptor: | C032 Fab Heavy Chain, C032 Fab Light Chain | | Authors: | Flyak, A.I, Bjorkman, P.J, Barnes, C.O. | | Deposit date: | 2021-06-01 | | Release date: | 2021-08-04 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Affinity maturation of SARS-CoV-2 neutralizing antibodies confers potency, breadth, and resilience to viral escape mutations.
Immunity, 54, 2021
|
|
7N3G
 
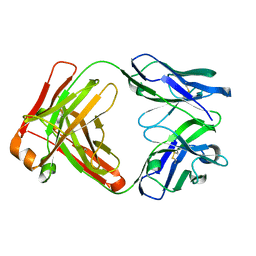 | | Crystal structure of an anti-SARS-CoV-2 human neutralizing antibody Fab fragment C098 | | Descriptor: | C098 Fab Heavy Chain, C098 Fab Light Chain | | Authors: | Flyak, A.I, Bjorkman, P.J, Barnes, C.O. | | Deposit date: | 2021-06-01 | | Release date: | 2021-08-04 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | Affinity maturation of SARS-CoV-2 neutralizing antibodies confers potency, breadth, and resilience to viral escape mutations.
Immunity, 54, 2021
|
|
7N3H
 
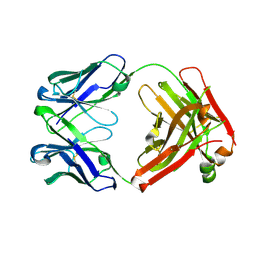 | | Crystal structure of an anti-SARS-CoV-2 human neutralizing antibody Fab fragment C099 | | Descriptor: | C099 Fab Heavy Chain, C099 Fab Light Chain | | Authors: | Flyak, A.I, Bjorkman, P.J, Barnes, C.O. | | Deposit date: | 2021-06-01 | | Release date: | 2021-08-04 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.26 Å) | | Cite: | Affinity maturation of SARS-CoV-2 neutralizing antibodies confers potency, breadth, and resilience to viral escape mutations.
Immunity, 54, 2021
|
|
7N3F
 
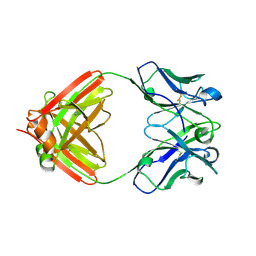 | | Crystal structure of an anti-SARS-CoV-2 human neutralizing antibody Fab fragment C080 | | Descriptor: | C080 Fab Heavy Chain, C080 Fab Light Chain | | Authors: | Flyak, A.I, Bjorkman, P.J, Barnes, C.O. | | Deposit date: | 2021-06-01 | | Release date: | 2021-08-04 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Affinity maturation of SARS-CoV-2 neutralizing antibodies confers potency, breadth, and resilience to viral escape mutations.
Immunity, 54, 2021
|
|
7N3I
 
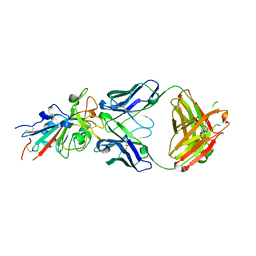 | | Crystal structure of the SARS-CoV-2 receptor binding domain in complex with the human neutralizing antibody Fab fragment C098 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, C098 Fab heavy chain, C098 Fab light chain, ... | | Authors: | Flyak, A.I, Bjorkman, P.J, Barnes, C.O. | | Deposit date: | 2021-06-01 | | Release date: | 2021-08-04 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Affinity maturation of SARS-CoV-2 neutralizing antibodies confers potency, breadth, and resilience to viral escape mutations.
Immunity, 54, 2021
|
|
7R8N
 
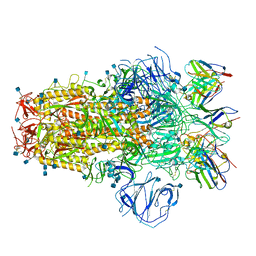 | | Structure of the SARS-CoV-2 S 6P trimer in complex with neutralizing antibody C051 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, C051 Fab Heavy Chain, ... | | Authors: | Barnes, C.O, Bjorkman, P.J. | | Deposit date: | 2021-06-26 | | Release date: | 2021-08-04 | | Last modified: | 2021-10-06 | | Method: | ELECTRON MICROSCOPY (3.55 Å) | | Cite: | Affinity maturation of SARS-CoV-2 neutralizing antibodies confers potency, breadth, and resilience to viral escape mutations.
Immunity, 54, 2021
|
|
7R8O
 
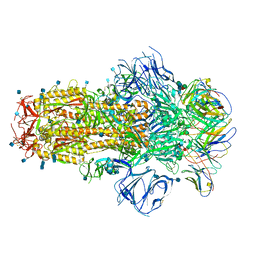 | | Structure of the SARS-CoV-2 S 6P trimer in complex with neutralizing antibody C548 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | DeLaitsch, A.T, Barnes, C.O, Bjorkman, P.J. | | Deposit date: | 2021-06-26 | | Release date: | 2021-08-04 | | Last modified: | 2021-10-13 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Affinity maturation of SARS-CoV-2 neutralizing antibodies confers potency, breadth, and resilience to viral escape mutations.
Immunity, 54, 2021
|
|
7R8L
 
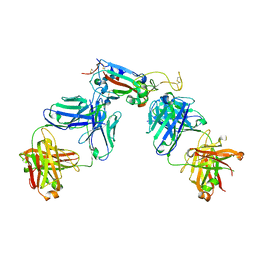 | | Structure of the SARS-CoV-2 RBD in complex with neutralizing antibody C099 and CR3022 | | Descriptor: | C099 Fab Heavy Chain, C099 Fab Light Chain, CR3022 Fab heavy chain, ... | | Authors: | Barnes, C.O, Bjorkman, P.J. | | Deposit date: | 2021-06-26 | | Release date: | 2021-08-04 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Affinity maturation of SARS-CoV-2 neutralizing antibodies confers potency, breadth, and resilience to viral escape mutations.
Immunity, 54, 2021
|
|
7R8M
 
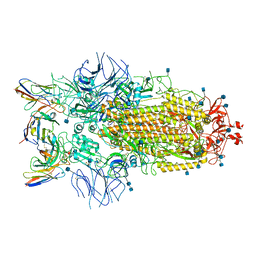 | | Structure of the SARS-CoV-2 S 6P trimer in complex with neutralizing antibody C032 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | DeLaitsch, A.T, Barnes, C.O, Bjorkman, P.J. | | Deposit date: | 2021-06-26 | | Release date: | 2021-08-04 | | Last modified: | 2021-10-06 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Affinity maturation of SARS-CoV-2 neutralizing antibodies confers potency, breadth, and resilience to viral escape mutations.
Immunity, 54, 2021
|
|
6XCM
 
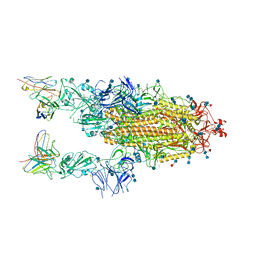 | |
6XCA
 
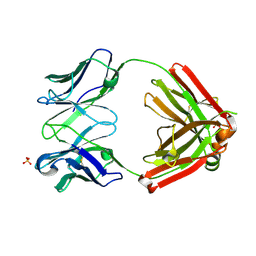 | | Crystal structure of an anti-SARS-CoV-2 human neutralizing antibody Fab fragment, C105 | | Descriptor: | C105 Heavy Chain, C105 Light Chain, SULFATE ION | | Authors: | Sharaf, N.G, Barnes, C.O, Bjorkman, P.J. | | Deposit date: | 2020-06-08 | | Release date: | 2020-07-01 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structures of Human Antibodies Bound to SARS-CoV-2 Spike Reveal Common Epitopes and Recurrent Features of Antibodies.
Cell, 182, 2020
|
|
6XCN
 
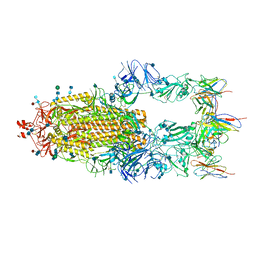 | |
7UCE
 
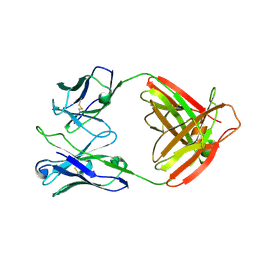 | |
7UCF
 
 | |
7UCG
 
 | |
5D4K
 
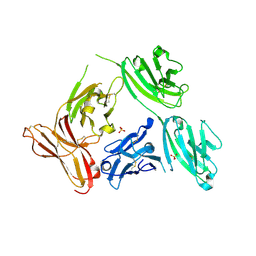 | |
5F1S
 
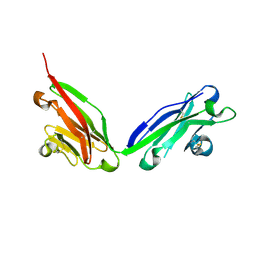 | |
7RKV
 
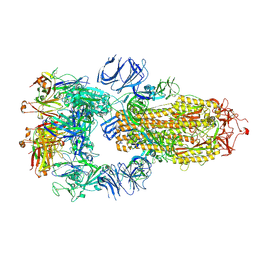 | | Structure of the SARS-CoV-2 S 6P trimer in complex with neutralizing antibody C118 (State 1) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, C118 Fab Heavy Chain, C118 Fab Light Chain, ... | | Authors: | Barnes, C.O, Jette, C.A, Bjorkman, P.J. | | Deposit date: | 2021-07-22 | | Release date: | 2021-09-22 | | Last modified: | 2021-10-13 | | Method: | ELECTRON MICROSCOPY (3.45 Å) | | Cite: | Broad cross-reactivity across sarbecoviruses exhibited by a subset of COVID-19 donor-derived neutralizing antibodies.
Cell Rep, 36, 2021
|
|
7RKU
 
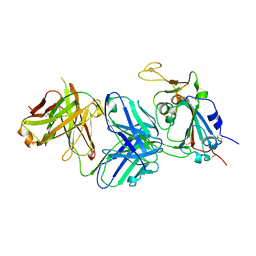 | | Structure of the SARS-CoV-2 receptor binding domain in complex with the human neutralizing antibody Fab fragment, C022 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, C022 Antibody Fab Heavy Chain, C022 Antibody Fab Light Chain, ... | | Authors: | Jette, C.A, Bjorkman, P.J, Barnes, C.O. | | Deposit date: | 2021-07-22 | | Release date: | 2021-09-22 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Broad cross-reactivity across sarbecoviruses exhibited by a subset of COVID-19 donor-derived neutralizing antibodies.
Cell Rep, 36, 2021
|
|
7RKS
 
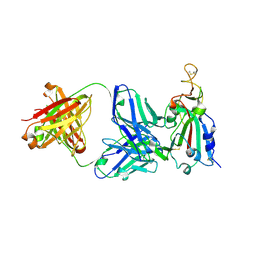 | | Structure of the SARS-CoV receptor binding domain in complex with the human neutralizing antibody Fab fragment, C118 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, C118 Antibody Fab Heavy Chain, ... | | Authors: | Jette, C.A, Bjorkman, P.J, Barnes, C.O. | | Deposit date: | 2021-07-22 | | Release date: | 2021-09-22 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Broad cross-reactivity across sarbecoviruses exhibited by a subset of COVID-19 donor-derived neutralizing antibodies.
Cell Rep, 36, 2021
|
|
7JG1
 
 | | Dimeric Immunoglobin A (dIgA) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Igh protein, ... | | Authors: | Kumar Bharathkar, S, Parker, B.P, Malyutin, A.G, Stadtmueller, B.M. | | Deposit date: | 2020-07-18 | | Release date: | 2020-11-11 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | The structures of Secretory and dimeric Immunoglobulin A.
Elife, 9, 2020
|
|
7JG2
 
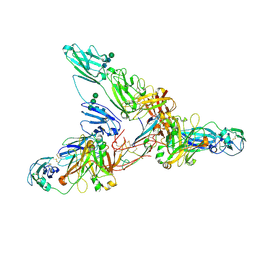 | | Secretory Immunoglobin A (SIgA) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Igh protein, ... | | Authors: | Kumar Bharathkar, S, Parker, B.P, Malyutin, A.G, Stadtmueller, B.M. | | Deposit date: | 2020-07-18 | | Release date: | 2020-11-11 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | The structures of Secretory and dimeric Immunoglobulin A.
Elife, 9, 2020
|
|
7K8N
 
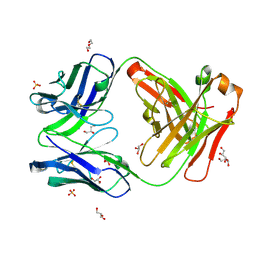 | | Crystal structure of an anti-SARS-CoV-2 human neutralizing antibody Fab fragment, C102 | | Descriptor: | C102 Fab Heavy Chain, C102 Fab Light Chain, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Jette, C.A, Barnes, C.O, Bjorkman, P.J. | | Deposit date: | 2020-09-27 | | Release date: | 2020-10-21 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | SARS-CoV-2 neutralizing antibody structures inform therapeutic strategies.
Nature, 588, 2020
|
|
