1DFN
 
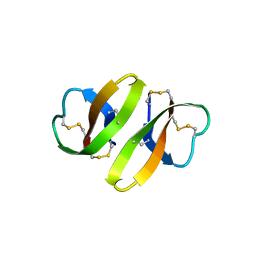 | | CRYSTAL STRUCTURE OF DEFENSIN HNP-3, AN AMPHIPHILIC DIMER: MECHANISMS OF MEMBRANE PERMEABILIZATION | | Descriptor: | DEFENSIN HNP-3 | | Authors: | Hill, C.P, Yee, J, Selsted, M.E, Eisenberg, D. | | Deposit date: | 1991-01-18 | | Release date: | 1992-07-15 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of defensin HNP-3, an amphiphilic dimer: mechanisms of membrane permeabilization.
Science, 251, 1991
|
|
1HIW
 
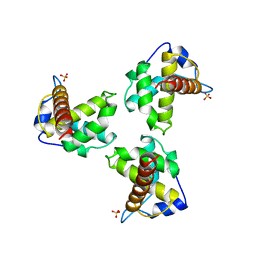 | | TRIMERIC HIV-1 MATRIX PROTEIN | | Descriptor: | HIV-1 MATRIX PROTEIN, SULFATE ION | | Authors: | Hill, C.P, Worthylake, D, Bancroft, D.P, Christensen, A.M, Sundquist, W.I. | | Deposit date: | 1996-02-28 | | Release date: | 1996-10-14 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structures of the trimeric human immunodeficiency virus type 1 matrix protein: implications for membrane association and assembly.
Proc.Natl.Acad.Sci.USA, 93, 1996
|
|
1AL1
 
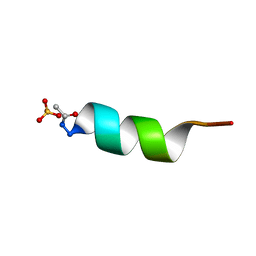 | | CRYSTAL STRUCTURE OF ALPHA1: IMPLICATIONS FOR PROTEIN DESIGN | | Descriptor: | ALPHA HELIX PEPTIDE: ELLKKLLEELKG, SULFATE ION | | Authors: | Hill, C.P, Anderson, D.H, Wesson, L, Degrado, W.F, Eisenberg, D. | | Deposit date: | 1990-07-02 | | Release date: | 1991-10-15 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of alpha 1: implications for protein design.
Science, 249, 1990
|
|
1RCM
 
 | |
1RHG
 
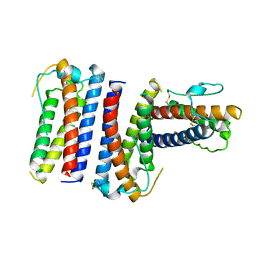 | |
4V7O
 
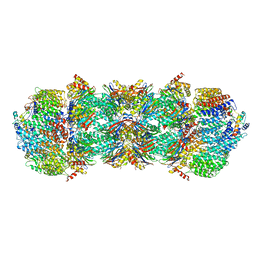 | | Proteasome Activator Complex | | Descriptor: | Proteasome activator BLM10, Proteasome component C1, Proteasome component C11, ... | | Authors: | Hill, C.P, Whitby, F.G. | | Deposit date: | 2009-12-22 | | Release date: | 2014-07-09 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3.005 Å) | | Cite: | Structure of a Blm10 complex reveals common mechanisms for proteasome binding and gate opening.
Mol.Cell, 37, 2010
|
|
7U2V
 
 | | Plasmodium falciparum Cyt c2 DSD | | Descriptor: | Cytochrome c2, HEME C | | Authors: | Hill, C.P, Wienkers, H.J, Whitby, F.G. | | Deposit date: | 2022-02-24 | | Release date: | 2023-05-10 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Direct tests of cytochrome c and c1 functions in the electron transport chain of malaria parasites
Proc Natl Acad Sci U S A, 120, 2023
|
|
6PSA
 
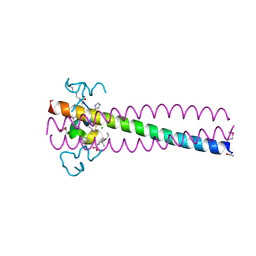 | | PIE12 D-PEPTIDE AGAINST HIV ENTRY (IN COMPLEX WITH IQN17 Q577R RESISTANCE MUTANT) | | Descriptor: | CHLORIDE ION, IQN17, PIE12 D-peptide | | Authors: | Hill, C.P, Whitby, F.G, Kay, M, Weinstock, M. | | Deposit date: | 2019-07-12 | | Release date: | 2020-02-05 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Characterization of resistance to a potent D-peptide HIV entry inhibitor.
Retrovirology, 16, 2019
|
|
7TXE
 
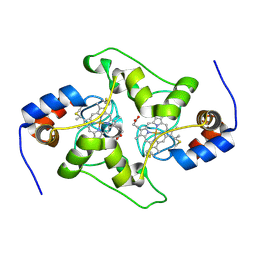 | | Plasmodium falciparum Cyt c2 DSD | | Descriptor: | Cytochrome c2, HEME C | | Authors: | Hill, C.P, Wienkers, H.J, Whitby, F.G. | | Deposit date: | 2022-02-08 | | Release date: | 2023-05-10 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Direct tests of cytochrome c and c1 functions in the electron transport chain of malaria parasites
Proc Natl Acad Sci U S A, 120, 2023
|
|
3GVW
 
 | | Single-chain UROD F217Y (YF) mutation | | Descriptor: | Uroporphyrinogen decarboxylase | | Authors: | Hill, C.P, Phillips, J.D, Whitby, F.G, Warby, C, Kushner, J.P. | | Deposit date: | 2009-03-31 | | Release date: | 2009-12-22 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Substrate shuttling between active sites of uroporphyrinogen decarboxylase is not required to generate coproporphyrinogen.
J.Mol.Biol., 389, 2009
|
|
3GVR
 
 | |
3GVV
 
 | | Single-chain UROD Y164G (GY) mutation | | Descriptor: | Uroporphyrinogen decarboxylase | | Authors: | Hill, C.P, Phillips, J.D, Whitby, F.G, Warby, C, Kushner, J.P. | | Deposit date: | 2009-03-31 | | Release date: | 2009-07-07 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Substrate shuttling between active sites of uroporphyrinogen decarboxylase is not required to generate coproporphyrinogen.
J.Mol.Biol., 389, 2009
|
|
3GW3
 
 | | human UROD mutant K297N | | Descriptor: | Uroporphyrinogen decarboxylase | | Authors: | Hill, C.P, Phillips, J.D, Whitby, F.G, Warby, C, Kushner, J.P. | | Deposit date: | 2009-03-31 | | Release date: | 2009-07-07 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural and kinetic characterization of mutant human uroporphyrinogen decarboxylases.
Cell Mol Biol (Noisy-le-grand), 55, 2009
|
|
3GVQ
 
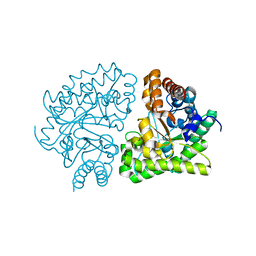 | | UROD single-chain dimer | | Descriptor: | Uroporphyrinogen decarboxylase | | Authors: | Hill, C.P, Phillips, J.D, Warby, C, Whitby, F.G, Kushner, J.P. | | Deposit date: | 2009-03-31 | | Release date: | 2009-07-07 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Substrate shuttling between active sites of uroporphyrinogen decarboxylase is not required to generate coproporphyrinogen.
J.Mol.Biol., 389, 2009
|
|
3GW0
 
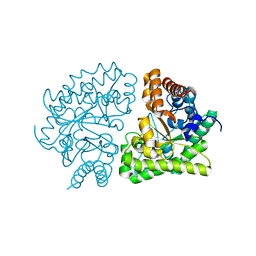 | | UROD mutant G318R | | Descriptor: | Uroporphyrinogen decarboxylase | | Authors: | Hill, C.P, Phillips, J.D, Whitby, F.G, Warby, C, Kushner, J.P. | | Deposit date: | 2009-03-31 | | Release date: | 2009-07-07 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural and kinetic characterization of mutant human uroporphyrinogen decarboxylases.
Cell Mol Biol (Noisy-le-grand), 55, 2009
|
|
3LL9
 
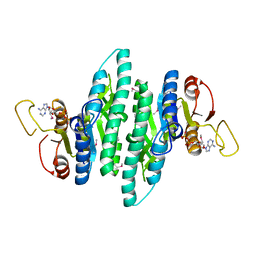 | |
3MGN
 
 | | D-Peptide inhibitor PIE71 in complex with IQN17 | | Descriptor: | D-PEPTIDE INHIBITOR PIE71, IQN17 | | Authors: | Hill, C.P, Whitby, F.G, Kay, M, Francis, N. | | Deposit date: | 2010-04-07 | | Release date: | 2011-03-02 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Design of a potent D-peptide HIV-1 entry inhibitor with a strong barrier to resistance.
J.Virol., 84, 2010
|
|
1YAB
 
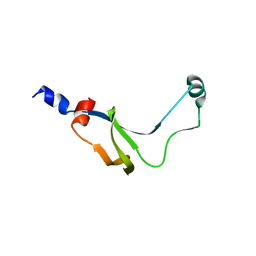 | | Structure of T. maritima FliN flagellar rotor protein | | Descriptor: | chemotaxis protein | | Authors: | Hill, C.P, Blair, D.F, Brown, P.N, Mathews, M.A.A, Joss, L.A. | | Deposit date: | 2004-12-17 | | Release date: | 2005-06-07 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Crystal Structure of the Flagellar Rotor Protein FliN from Thermotoga maritima
J.BACTERIOL., 187, 2005
|
|
3FRV
 
 | |
1QB8
 
 | |
1AK4
 
 | | HUMAN CYCLOPHILIN A BOUND TO THE AMINO-TERMINAL DOMAIN OF HIV-1 CAPSID | | Descriptor: | CYCLOPHILIN A, HIV-1 CAPSID | | Authors: | Hill, C.P, Gamble, T.R, Vajdos, F.F, Worthylake, D.K, Sundquist, W.I. | | Deposit date: | 1997-05-28 | | Release date: | 1997-10-15 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.36 Å) | | Cite: | Crystal structure of human cyclophilin A bound to the amino-terminal domain of HIV-1 capsid.
Cell(Cambridge,Mass.), 87, 1996
|
|
1AVO
 
 | | PROTEASOME ACTIVATOR REG(ALPHA) | | Descriptor: | 11S REGULATOR | | Authors: | Hill, C.P, Knowlton, J.R. | | Deposit date: | 1997-09-18 | | Release date: | 1997-12-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure of the proteasome activator REGalpha (PA28alpha).
Nature, 390, 1997
|
|
1AUM
 
 | | HIV CAPSID C-TERMINAL DOMAIN (CAC146) | | Descriptor: | HIV CAPSID | | Authors: | Hill, C.P, Gamble, T.R, Yoo, S, Vajdos, F.F, Von Schwedler, U.K, Worthylake, D.K, Wang, H, Mccutcheon, J.P, Sundquist, W.I. | | Deposit date: | 1997-08-29 | | Release date: | 1998-01-14 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structure of the carboxyl-terminal dimerization domain of the HIV-1 capsid protein.
Science, 278, 1997
|
|
2R03
 
 | |
2R05
 
 | |
