4DW0
 
 | |
4DW1
 
 | |
3A1U
 
 | | Crystal structue of the cytosolic domain of T. maritima FeoB iron iransporter in GMPPNP form | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, Iron(II) transport protein B, MAGNESIUM ION, ... | | Authors: | Hattori, M, Ishitani, R, Nureki, O. | | Deposit date: | 2009-04-22 | | Release date: | 2009-09-22 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural basis of novel interactions between the small-GTPase and GDI-like domains in prokaryotic FeoB iron transporter
Structure, 17, 2009
|
|
3A1S
 
 | | Crystal structue of the cytosolic domain of T. maritima FeoB iron iransporter in GDP form I | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Hattori, M, Ishitani, R, Nureki, O. | | Deposit date: | 2009-04-22 | | Release date: | 2009-09-22 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural basis of novel interactions between the small-GTPase and GDI-like domains in prokaryotic FeoB iron transporter
Structure, 17, 2009
|
|
3A1W
 
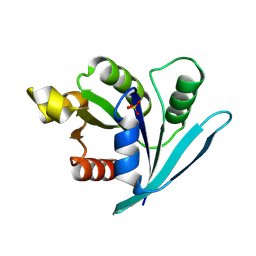 | |
3A1T
 
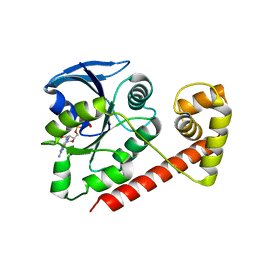 | |
3A1V
 
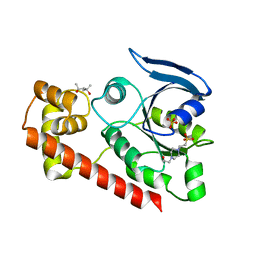 | | Crystal structue of the cytosolic domain of T. maritima FeoB iron iransporter in apo form | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ... | | Authors: | Hattori, M, Ishitani, R, Nureki, O. | | Deposit date: | 2009-04-22 | | Release date: | 2009-09-22 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural basis of novel interactions between the small-GTPase and GDI-like domains in prokaryotic FeoB iron transporter
Structure, 17, 2009
|
|
2YVZ
 
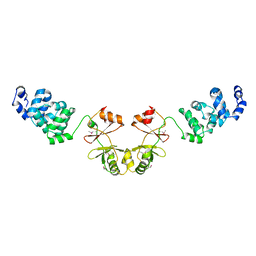 | | Crystal structure of magnesium transporter MgtE cytosolic domain, Mg2+-free form | | Descriptor: | Mg2+ transporter MgtE | | Authors: | Hattori, M, Tanaka, Y, Fukai, S, Ishitani, R, Nureki, O. | | Deposit date: | 2007-04-18 | | Release date: | 2007-09-04 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (3.9 Å) | | Cite: | Crystal structure of the MgtE Mg(2+) transporter
Nature, 448, 2007
|
|
2YVY
 
 | | Crystal structure of magnesium transporter MgtE cytosolic domain, Mg2+ bound form | | Descriptor: | MAGNESIUM ION, Mg2+ transporter MgtE | | Authors: | Hattori, M, Tanaka, Y, Fukai, S, Ishitani, R, Nureki, O. | | Deposit date: | 2007-04-18 | | Release date: | 2007-09-04 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of the MgtE Mg(2+) transporter
Nature, 448, 2007
|
|
2YVX
 
 | | Crystal structure of magnesium transporter MgtE | | Descriptor: | MAGNESIUM ION, Mg2+ transporter MgtE | | Authors: | Hattori, M, Tanaka, Y, Fukai, S, Ishitani, R, Nureki, O. | | Deposit date: | 2007-04-18 | | Release date: | 2007-09-04 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Crystal structure of the MgtE Mg(2+) transporter
Nature, 448, 2007
|
|
8JV5
 
 | |
8JV6
 
 | |
6AH5
 
 | | Structure of human P2X3 receptor in complex with ATP and Mg2+ ion | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ADENOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Hattori, M. | | Deposit date: | 2018-08-16 | | Release date: | 2019-06-12 | | Last modified: | 2020-12-23 | | Method: | X-RAY DIFFRACTION (3.819 Å) | | Cite: | Molecular mechanisms of human P2X3 receptor channel activation and modulation by divalent cation bound ATP.
Elife, 8, 2019
|
|
6AH4
 
 | | Structure of human P2X3 receptor in complex with ATP and Ca2+ ion | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ADENOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Hattori, M. | | Deposit date: | 2018-08-16 | | Release date: | 2019-06-12 | | Last modified: | 2020-12-23 | | Method: | X-RAY DIFFRACTION (3.296 Å) | | Cite: | Molecular mechanisms of human P2X3 receptor channel activation and modulation by divalent cation bound ATP.
Elife, 8, 2019
|
|
1WV8
 
 | | Crystal structure of hypothetical protein TTHA1013 from an extremely thermophilic bacterium thermus thermophilus HB8 | | Descriptor: | hypothetical protein TTHA1013 | | Authors: | Mizohata, E, Hattori, M, Kuramitsu, S, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2004-12-12 | | Release date: | 2005-06-12 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of the hypothetical protein TTHA1013 from Thermus thermophilus HB8
Proteins, 61, 2005
|
|
1WV9
 
 | | Crystal Structure of Rhodanese Homolog TT1651 from an Extremely Thermophilic Bacterium Thermus thermophilus HB8 | | Descriptor: | Rhodanese Homolog TT1651 | | Authors: | Mizohata, E, Hattori, M, Tatsuguchi, A, Terada, T, Kuramitsu, S, Shirouzu, M, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2004-12-12 | | Release date: | 2005-06-19 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of the single-domain rhodanese homologue TTHA0613 from Thermus thermophilus HB8
Proteins, 64, 2006
|
|
6LBH
 
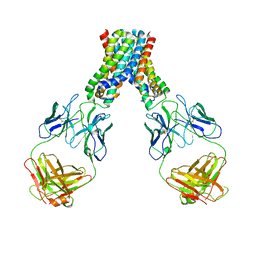 | |
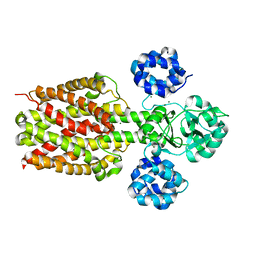
2ZY9
 
 | |
5X9H
 
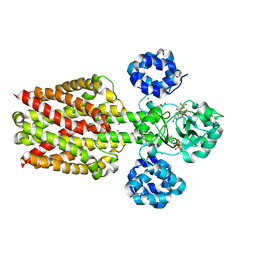 | |
5X9G
 
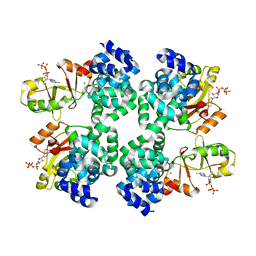 | |
8H5E
 
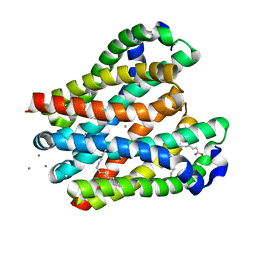 | | Crystal structure of the MgtE TM domain in complex with Ca2+ ions at 2.5 angstrom resolution | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, CALCIUM ION, Magnesium transporter MgtE | | Authors: | Teng, X, Sheng, D, Hattori, M. | | Deposit date: | 2022-10-13 | | Release date: | 2022-12-07 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Ion selectivity mechanism of the MgtE channel for Mg 2+ over Ca 2 .
Iscience, 25, 2022
|
|
8H70
 
 | |
8JV7
 
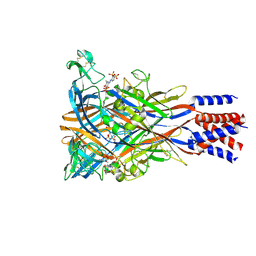 | | Cryo-EM structure of the panda P2X7 receptor in complex with PPADS | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 4-[(E)-[4-methanoyl-6-methyl-5-oxidanyl-3-(phosphonooxymethyl)pyridin-2-yl]diazenyl]benzene-1,3-disulfonic acid, P2X purinoceptor | | Authors: | Sheng, D, Hattori, M. | | Deposit date: | 2023-06-27 | | Release date: | 2023-11-29 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Structural insights into the orthosteric inhibition of P2X receptors by non-ATP analog antagonists.
Elife, 12, 2024
|
|
8JV8
 
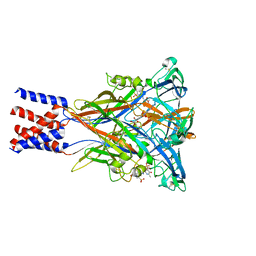 | | Cryo-EM structure of the panda P2X7 receptor in complex with PPNDS | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 3-[(E)-{4-formyl-5-hydroxy-6-methyl-3-[(phosphonooxy)methyl]pyridin-2-yl}diazenyl]-7-nitronaphthalene-1,5-disulfonic acid, P2X purinoceptor | | Authors: | Sheng, D, Hattori, M. | | Deposit date: | 2023-06-27 | | Release date: | 2023-11-29 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.34 Å) | | Cite: | Structural insights into the orthosteric inhibition of P2X receptors by non-ATP analog antagonists.
Elife, 12, 2024
|
|
5WZY
 
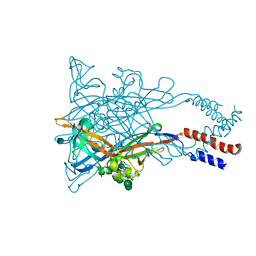 | | Crystal structure of the P2X4 receptor from zebrafish in the presence of CTP at 2.8 Angstroms | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CYTIDINE-5'-TRIPHOSPHATE, GLYCEROL, ... | | Authors: | Kasuya, G, Hattori, M, Nureki, O. | | Deposit date: | 2017-01-19 | | Release date: | 2017-04-05 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.799 Å) | | Cite: | Structural insights into the nucleotide base specificity of P2X receptors
Sci Rep, 7, 2017
|
|
