7UCR
 
 | | Joint X-ray/neutron structure of the Sarcin-Ricin loop RNA | | Descriptor: | SULFATE ION, Sarcin-Ricin loop RNA | | Authors: | Harp, J.M, Egli, M.E, Pallan, P.S, Coates, L. | | Deposit date: | 2022-03-17 | | Release date: | 2022-07-20 | | Last modified: | 2024-04-03 | | Method: | NEUTRON DIFFRACTION (1 Å), X-RAY DIFFRACTION | | Cite: | Cryo neutron crystallography demonstrates influence of RNA 2'-OH orientation on conformation, sugar pucker and water structure.
Nucleic Acids Res., 50, 2022
|
|
7JY2
 
 | | Z-DNA joint X-ray/Neutron | | Descriptor: | Chains: A,B | | Authors: | Harp, J.M, Coates, L, Egli, M. | | Deposit date: | 2020-08-28 | | Release date: | 2021-04-28 | | Last modified: | 2024-04-03 | | Method: | NEUTRON DIFFRACTION (1.5 Å), X-RAY DIFFRACTION | | Cite: | Water structure around a left-handed Z-DNA fragment analyzed by cryo neutron crystallography.
Nucleic Acids Res., 49, 2021
|
|
6CXZ
 
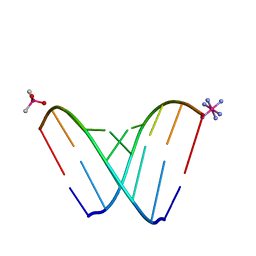 | | RNA octamer containing 2'-F, 4'-Calpha-Me U. | | Descriptor: | CACODYLATE ION, COBALT HEXAMMINE(III), RNA (5'-R(*CP*GP*AP*AP*(U4M)P*UP*CP*G)-3') | | Authors: | Harp, J.M, Egli, M. | | Deposit date: | 2018-04-04 | | Release date: | 2018-08-29 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural basis for the synergy of 4'- and 2'-modifications on siRNA nuclease resistance, thermal stability and RNAi activity.
Nucleic Acids Res., 46, 2018
|
|
6CY0
 
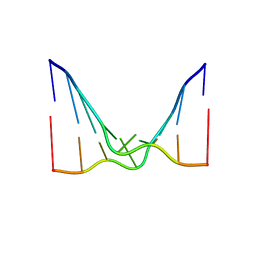 | | RNA octamer containing 2'-F, 4'-Cbeta-OMe U. | | Descriptor: | RNA (5'-R(*(CBV)P*GP*AP*AP*(UFB)P*UP*CP*G)-3') | | Authors: | Harp, J.M, Egli, M. | | Deposit date: | 2018-04-04 | | Release date: | 2018-08-29 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.398 Å) | | Cite: | Structural basis for the synergy of 4'- and 2'-modifications on siRNA nuclease resistance, thermal stability and RNAi activity.
Nucleic Acids Res., 46, 2018
|
|
6CY2
 
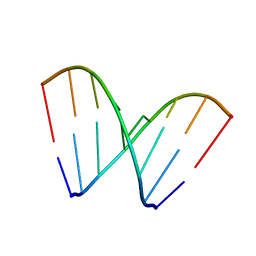 | | RNA octamer containing 2'-OMe, 4'Calpha-OMe U. | | Descriptor: | RNA (5'-R(*(CBV)P*GP*AP*AP*(UOA)P*UP*CP*G)-3') | | Authors: | Harp, J.M, Egli, M. | | Deposit date: | 2018-04-04 | | Release date: | 2018-08-29 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structural basis for the synergy of 4'- and 2'-modifications on siRNA nuclease resistance, thermal stability and RNAi activity.
Nucleic Acids Res., 46, 2018
|
|
6CY4
 
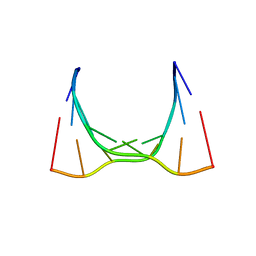 | | RNA octamer containing 2'-OMe, 4'- Cbeta-OMe U. | | Descriptor: | RNA (5'-R(*(CBV)P*GP*AP*AP*(UOB)P*UP*CP*G)-3') | | Authors: | Harp, J.M, Egli, M. | | Deposit date: | 2018-04-04 | | Release date: | 2018-08-29 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.851 Å) | | Cite: | Structural basis for the synergy of 4'- and 2'-modifications on siRNA nuclease resistance, thermal stability and RNAi activity.
Nucleic Acids Res., 46, 2018
|
|
9BF0
 
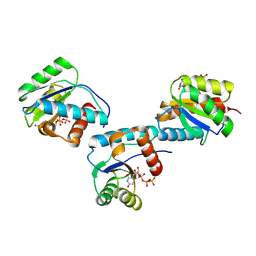 | | MID domain of human Argo2 bound to UTP | | Descriptor: | Protein argonaute-2, URIDINE 5'-TRIPHOSPHATE | | Authors: | Harp, J.M, Egli, M. | | Deposit date: | 2024-04-16 | | Release date: | 2024-07-10 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Structure and Stability of Ago2 MID-Nucleotide Complexes: All-in-One (Drop) His 6 -SUMO Tag Removal, Nucleotide Binding, and Crystal Growth.
Curr Protoc, 4, 2024
|
|
9BF2
 
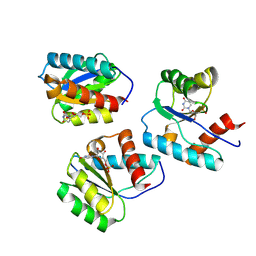 | | MID domain of Ago2 bound to UMP | | Descriptor: | Protein argonaute-2, URIDINE-5'-MONOPHOSPHATE | | Authors: | Harp, J.M, Egli, M. | | Deposit date: | 2024-04-16 | | Release date: | 2024-07-10 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Structure and Stability of Ago2 MID-Nucleotide Complexes: All-in-One (Drop) His 6 -SUMO Tag Removal, Nucleotide Binding, and Crystal Growth.
Curr Protoc, 4, 2024
|
|
9BEZ
 
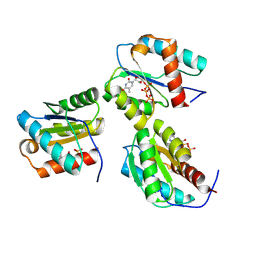 | | MID domain of human Argo2 bound to RNA | | Descriptor: | Protein argonaute-2, [(3~{S},4~{R},5~{R})-5-[5-methyl-2,4-bis(oxidanylidene)pyrimidin-1-yl]-4-oxidanyl-oxolan-3-yl] [oxidanyl(phosphonooxy)phosphoryl] hydrogen phosphate | | Authors: | Harp, J.M, Egli, M. | | Deposit date: | 2024-04-16 | | Release date: | 2024-07-10 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure and Stability of Ago2 MID-Nucleotide Complexes: All-in-One (Drop) His 6 -SUMO Tag Removal, Nucleotide Binding, and Crystal Growth.
Curr Protoc, 4, 2024
|
|
1EQZ
 
 | | X-RAY STRUCTURE OF THE NUCLEOSOME CORE PARTICLE AT 2.5 A RESOLUTION | | Descriptor: | 146 NUCLEOTIDES LONG DNA, CACODYLATE ION, CHLORIDE ION, ... | | Authors: | Hanson, B.L, Harp, J.M, Timm, D.E, Bunick, G.J. | | Deposit date: | 2000-04-06 | | Release date: | 2000-04-17 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Asymmetries in the nucleosome core particle at 2.5 A resolution.
Acta Crystallogr.,Sect.D, 56, 2000
|
|
7JJF
 
 | | Sarcin-ricin loop with modified residue. | | Descriptor: | MAGNESIUM ION, RNA/DNA (27-mer) | | Authors: | Harp, J.M, Pallan, P.S, Egli, M. | | Deposit date: | 2020-07-25 | | Release date: | 2020-12-23 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Incorporating a Thiophosphate Modification into a Common RNA Tetraloop Motif Causes an Unanticipated Stability Boost.
Biochemistry, 59, 2020
|
|
7LO9
 
 | | RNA dodecamer containing a GNA A residue | | Descriptor: | Chains: A,B,C,D | | Authors: | Harp, J.M, Wawrzak, Z, Egli, M. | | Deposit date: | 2021-02-09 | | Release date: | 2021-12-22 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Overcoming GNA/RNA base-pairing limitations using isonucleotides improves the pharmacodynamic activity of ESC+ GalNAc-siRNAs.
Nucleic Acids Res., 49, 2021
|
|
8SKQ
 
 | |
9O9N
 
 | | Crystal structure of PPARgamma ligand binding domain (LBD) in complex with NCoR1 corepressor peptide and inverse agonist FX-909 | | Descriptor: | (3P)-3-(5,7-difluoro-4-oxo-1,4-dihydroquinolin-2-yl)-4-(methanesulfonyl)benzonitrile, Nuclear receptor corepressor 1, Peroxisome proliferator-activated receptor gamma | | Authors: | Laughlin, Z.T, Dong, J, Harp, J.M, Kojetin, D.J. | | Deposit date: | 2025-04-18 | | Release date: | 2025-05-14 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural basis of PPARgamma-mediated transcriptional repression by the covalent inverse agonist FX-909
To Be Published
|
|
3MFA
 
 | | Computationally designed endo-1,4-beta-xylanase | | Descriptor: | Endo-1,4-beta-xylanase, SULFATE ION | | Authors: | Morin, A, Harp, J.M. | | Deposit date: | 2010-04-01 | | Release date: | 2010-11-10 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Computational design of an endo-1,4-{beta}-xylanase ligand binding site.
Protein Eng.Des.Sel., 24, 2011
|
|
3MF9
 
 | | Computationally designed endo-1,4-beta-xylanase | | Descriptor: | Endo-1,4-beta-xylanase, SULFATE ION | | Authors: | Morin, A, Harp, J.M. | | Deposit date: | 2010-04-01 | | Release date: | 2010-11-10 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Computational design of an endo-1,4-{beta}-xylanase ligand binding site.
Protein Eng.Des.Sel., 24, 2011
|
|
3MFC
 
 | | Computationally designed end0-1,4-beta,xylanase | | Descriptor: | Endo-1,4-beta-xylanase, SULFATE ION | | Authors: | Morin, A, Harp, J.M. | | Deposit date: | 2010-04-01 | | Release date: | 2010-11-10 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Computational design of an endo-1,4-{beta}-xylanase ligand binding site.
Protein Eng.Des.Sel., 24, 2011
|
|
3MF6
 
 | | Computationally designed endo-1,4-beta-xylanase | | Descriptor: | Endo-1,4-beta-xylanase, SULFATE ION | | Authors: | Morin, A, Harp, J.M. | | Deposit date: | 2010-04-01 | | Release date: | 2010-11-10 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.28 Å) | | Cite: | Computational design of an endo-1,4-{beta}-xylanase ligand binding site.
Protein Eng.Des.Sel., 24, 2011
|
|
8FTJ
 
 | | Crystal structure of human NEIL1 (P2G (242K) C(delta)100) glycosylase bound to DNA duplex containing urea | | Descriptor: | 1,2-ETHANEDIOL, DNA (5'-D(*CP*GP*TP*CP*CP*AP*UDV*GP*TP*CP*TP*AP*CP)-3'), DNA (5'-D(*TP*AP*GP*AP*CP*AP*TP*GP*GP*AP*CP*GP*G)-3'), ... | | Authors: | Tomar, R, Sharma, P, Harp, J.M, Egli, M, Stone, M.P. | | Deposit date: | 2023-01-12 | | Release date: | 2023-04-26 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Base excision repair of the N-(2-deoxy-d-erythro-pentofuranosyl)-urea lesion by the hNEIL1 glycosylase.
Nucleic Acids Res., 51, 2023
|
|
3R3E
 
 | | The glutathione bound structure of YqjG, a glutathione transferase homolog from Escherichia coli K-12 | | Descriptor: | GLUTATHIONE, SULFATE ION, Uncharacterized protein yqjG | | Authors: | Branch, M.C, Cook, P.D, Harp, J.M, Armstrong, R.N. | | Deposit date: | 2011-03-15 | | Release date: | 2012-03-14 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.205 Å) | | Cite: | Crystal Structure Analysis of yqjG from Escherichia Coli K-12
To be Published
|
|
7JJE
 
 | |
7JJD
 
 | |
1MT6
 
 | | Structure of histone H3 K4-specific methyltransferase SET7/9 with AdoHcy | | Descriptor: | S-ADENOSYL-L-HOMOCYSTEINE, SET9 | | Authors: | Jacobs, S.A, Harp, J.M, Devarakonda, S, Kim, Y, Rastinejad, F, Khorasanizadeh, S. | | Deposit date: | 2002-09-20 | | Release date: | 2002-11-06 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The active site of the SET domain is constructed on a knot
Nat.Struct.Biol., 9, 2002
|
|
1MUF
 
 | | Structure of histone H3 K4-specific methyltransferase SET7/9 | | Descriptor: | SET9 | | Authors: | Jacobs, S.A, Harp, J.M, Devarakonda, S, Kim, Y, Rastinejad, F, Khorasanizadeh, S. | | Deposit date: | 2002-09-23 | | Release date: | 2002-11-06 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | The active site of the SET domain is constructed on a knot
Nat.Struct.Biol., 9, 2002
|
|
7L5M
 
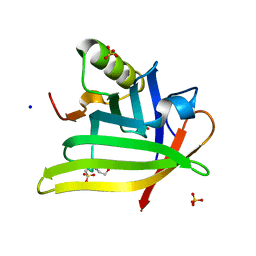 | | Crystal Structure of the DiB-RM-split Protein | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Lipocalin family protein, SODIUM ION, ... | | Authors: | Bozhanova, N.G, Harp, J.M, Meiler, J. | | Deposit date: | 2020-12-22 | | Release date: | 2021-10-27 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.33 Å) | | Cite: | Computational redesign of a fluorogen activating protein with Rosetta.
Plos Comput.Biol., 17, 2021
|
|
