3PQC
 
 | | Crystal structure of Thermotoga maritima ribosome biogenesis GTP-binding protein EngB (YsxC/YihA) in complex with GDP | | Descriptor: | DI(HYDROXYETHYL)ETHER, GUANOSINE-5'-DIPHOSPHATE, Probable GTP-binding protein engB | | Authors: | Chan, K.H, Wong, K.B. | | Deposit date: | 2010-11-26 | | Release date: | 2011-06-08 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure of an essential GTPase, YsxC, from Thermotoga maritima
Acta Crystallogr.,Sect.F, 67, 2011
|
|
3PR1
 
 | |
9ATW
 
 | | Structure of biofilm-forming functional amyloid PSMa1 from Staphylococcus aureus | | Descriptor: | Phenol-soluble modulin alpha 1 peptide | | Authors: | Hansen, K.H, Byeon, C.H, Liu, Q, Drace, T, Boesen, T, Conway, J.F, Andreasen, M, Akbey, U. | | Deposit date: | 2024-02-27 | | Release date: | 2024-08-07 | | Last modified: | 2024-08-21 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structure of biofilm-forming functional amyloid PSM alpha 1 from Staphylococcus aureus.
Proc.Natl.Acad.Sci.USA, 121, 2024
|
|
2YLO
 
 | | TARGETING THE BINDING FUNCTION 3 SITE OF THE ANDROGEN RECEPTOR THROUGH IN SILICO MOLECULAR MODELING | | Descriptor: | 1-[2-(4-METHYLPHENOXY)ETHYL]-2-(2-PHENOXYETHYLSULFANYL)BENZIMIDAZOLE, ANDROGEN RECEPTOR, SULFATE ION, ... | | Authors: | Lack, N.A, Axerio, P, Tavassoli, P, Kuchenbecker, K, Han, F.Q, Chan, K.H, Feau, C, LeBlanc, E, Tomlinson, E, Guy, R.K, Rennie, P.S, Cherkasov, A. | | Deposit date: | 2011-06-04 | | Release date: | 2011-07-06 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Targeting the Binding Function 3 (Bf3) Site of the Human Androgen Receptor Through Virtual Screening.
J.Med.Chem., 54, 2011
|
|
2YLP
 
 | | TARGETING THE BINDING FUNCTION 3 SITE OF THE ANDROGEN RECEPTOR THROUGH IN SILICO MOLECULAR MODELING | | Descriptor: | 3-[(2,4-DICHLOROPHENYL)METHYLSULFANYLMETHYL]BENZOIC ACID, ANDROGEN RECEPTOR, SULFATE ION, ... | | Authors: | Lack, N.A, Axerio, P, Tavassoli, P, Kuchenbecker, K, Han, F.Q, Chan, K.H, Feau, C, LeBlanc, E, Tomlinson, E, Guy, R.K, Rennie, P.S, Cherkasov, A. | | Deposit date: | 2011-06-04 | | Release date: | 2011-07-06 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Targeting the Binding Function 3 (Bf3) Site of the Human Androgen Receptor Through Virtual Screening.
J.Med.Chem., 54, 2011
|
|
2YLQ
 
 | | TARGETING THE BINDING FUNCTION 3 SITE OF THE ANDROGEN RECEPTOR THROUGH IN SILICO MOLECULAR MODELING | | Descriptor: | 3-[1-[2-(4-METHYLPHENOXY)ETHYL]BENZIMIDAZOL-2-YL]SULFANYLPROPANOIC ACID, ANDROGEN RECEPTOR, SULFATE ION, ... | | Authors: | Lack, N.A, Axerio, P, Tavassoli, P, Kuchenbecker, K, Han, F.Q, Chan, K.H, Feau, C, LeBlanc, E, Tomlinson, E, Guy, R.K, Rennie, P.S, Cherkasov, A. | | Deposit date: | 2011-06-04 | | Release date: | 2011-07-06 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Targeting the Binding Function 3 (Bf3) Site of the Human Androgen Receptor Through Virtual Screening.
J.Med.Chem., 54, 2011
|
|
1WZ4
 
 | |
3ZQT
 
 | | TARGETING THE BINDING FUNCTION 3 SITE OF THE ANDROGEN RECEPTOR THROUGH IN SILICO MOLECULAR MODELING | | Descriptor: | 4-[(2R,3S)-3-[(3,4-DIHYDROXYPHENYL)METHYL]-2-METHYLBUTYL]BENZENE-1,2-DIOL, ANDROGEN RECEPTOR, SULFATE ION, ... | | Authors: | Lack, N.A, Axerio, P, Tavassoli, P, Kuchenbecker, K, Han, F.Q, Chan, K.H, Feau, C, LeBlanc, E, Tomlinson, E, Guy, R.K, Rennie, P.S, Cherkasov, A. | | Deposit date: | 2011-06-10 | | Release date: | 2011-12-07 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | Targeting the Binding Function 3 (Bf3) Site of the Human Androgen Receptor Through Virtual Screening.
J.Med.Chem., 54, 2011
|
|
1K64
 
 | | NMR Structue of alpha-conotoxin EI | | Descriptor: | alpha-conotoxin EI | | Authors: | Park, K.H, Suk, J.E, Jacobsen, R, Gray, W.R, McIntosh, J.M, Han, K.H. | | Deposit date: | 2001-10-15 | | Release date: | 2003-09-09 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | Solution conformation of alpha-conotoxin EI, a neuromuscular toxin specific for the alpha 1/delta subunit interface of torpedo nicotinic acetylcholine receptor
J.BIOL.CHEM., 276, 2001
|
|
1K1D
 
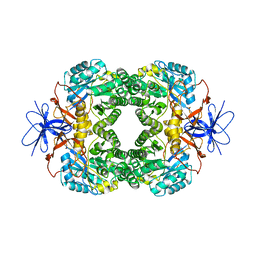 | | Crystal structure of D-hydantoinase | | Descriptor: | D-hydantoinase, ZINC ION | | Authors: | Cheon, Y.H, Kim, H.S, Han, K.H, Abendroth, J, Niefind, K, Schomburg, D, Wang, J, Kim, Y. | | Deposit date: | 2001-09-25 | | Release date: | 2002-08-14 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (3.01 Å) | | Cite: | Crystal structure of D-hydantoinase from Bacillus stearothermophilus: insight into the stereochemistry of enantioselectivity.
Biochemistry, 41, 2002
|
|
2YHD
 
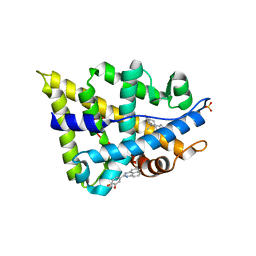 | | Human androgen receptor in complex with AF2 small molecule inhibitor | | Descriptor: | 4-(2,3-dihydro-1H-perimidin-2-yl)benzene-1,2-diol, ANDROGEN RECEPTOR, SULFATE ION, ... | | Authors: | AxerioCilies, P, Lack, N.A, ShashiNayana, M.R, Chan, K.H, Yeung, A, LeBlanc, E, Guns, E, Rennie, P, Cherkasov, A. | | Deposit date: | 2011-04-28 | | Release date: | 2011-09-21 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Inhibitors of Androgen Receptor Activation Function-2 (Af2) Site Identified Through Virtual Screening.
J.Med.Chem., 54, 2011
|
|
1P1P
 
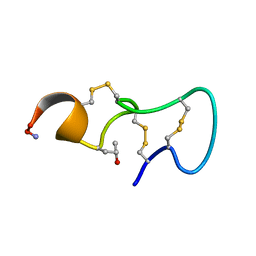 | | [PRO7,13] AA-CONOTOXIN PIVA, NMR, 12 STRUCTURES | | Descriptor: | AA-CONOTOXIN PIVA | | Authors: | Han, K.-H, Hwang, K.-J, Kim, S.-M, Kim, S.-K, Gray, W.R, Olivera, B.M, Rivier, J, Shon, K.J. | | Deposit date: | 1996-12-06 | | Release date: | 1997-07-07 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | NMR structure determination of a novel conotoxin, [Pro 7,13] alpha A-conotoxin PIVA.
Biochemistry, 36, 1997
|
|
6YSR
 
 | | Structure of the P+9 stalled ribosome complex | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Chan, K.-H, Petrychenko, V, Mueller, C, Maracci, C, Holtkamp, W, Wilson, D.N, Fischer, N, Rodnina, M.V. | | Deposit date: | 2020-04-23 | | Release date: | 2020-08-19 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Mechanism of ribosome rescue by alternative ribosome-rescue factor B.
Nat Commun, 11, 2020
|
|
6YSU
 
 | | Structure of the P+0 ArfB-ribosome complex in the post-hydrolysis state | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Chan, K.-H, Petrychenko, V, Mueller, C, Maracci, C, Holtkamp, W, Wilson, D.N, Fischer, N, Rodnina, M.V. | | Deposit date: | 2020-04-23 | | Release date: | 2020-08-19 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Mechanism of ribosome rescue by alternative ribosome-rescue factor B.
Nat Commun, 11, 2020
|
|
6YSS
 
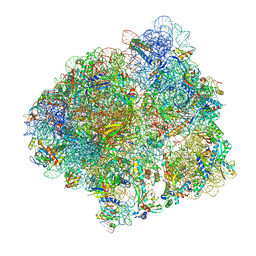 | | Structure of the P+9 ArfB-ribosome complex in the post-hydrolysis state | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Chan, K.-H, Petrychenko, V, Mueller, C, Maracci, C, Holtkamp, W, Wilson, D.N, Fischer, N, Rodnina, M.V. | | Deposit date: | 2020-04-23 | | Release date: | 2020-08-19 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Mechanism of ribosome rescue by alternative ribosome-rescue factor B.
Nat Commun, 11, 2020
|
|
6YST
 
 | | Structure of the P+9 ArfB-ribosome complex with P/E hybrid tRNA in the post-hydrolysis state | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Chan, K.-H, Petrychenko, V, Mueller, C, Maracci, C, Holtkamp, W, Wilson, D.N, Fischer, N, Rodnina, M.V. | | Deposit date: | 2020-04-23 | | Release date: | 2020-08-19 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Mechanism of ribosome rescue by alternative ribosome-rescue factor B.
Nat Commun, 11, 2020
|
|
7T32
 
 | | CryoEM structure of the adenosine 2A receptor-BRIL/Anti BRIL Fab complex with ZM241385 | | Descriptor: | 4-{2-[(7-amino-2-furan-2-yl[1,2,4]triazolo[1,5-a][1,3,5]triazin-5-yl)amino]ethyl}phenol, Adenosine receptor A2a/Soluble cytochrome b562 Fusion Protein | | Authors: | Zhang, K.H, Wu, H, Hoppe, N, Manglik, A, Cheng, Y.F. | | Deposit date: | 2021-12-06 | | Release date: | 2022-08-10 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Fusion protein strategies for cryo-EM study of G protein-coupled receptors.
Nat Commun, 13, 2022
|
|
2WSM
 
 | |
1QS3
 
 | | NMR SOLUTION CONFORMATION OF AN ANTITOXIC ANALOG OF ALPHA-CONOTOXIN GI | | Descriptor: | DES-GLU1-[CYS3ALA]-DES-CYS13-ALPHA CONOTOXIN GI | | Authors: | Mok, K.H, Han, K.-H. | | Deposit date: | 1999-06-25 | | Release date: | 1999-10-06 | | Last modified: | 2014-03-12 | | Method: | SOLUTION NMR | | Cite: | NMR solution conformation of an antitoxic analogue of alpha-conotoxin GI: identification of a common nicotinic acetylcholine receptor alpha 1-subunit binding surface for small ligands and alpha-conotoxins.
Biochemistry, 38, 1999
|
|
4MHJ
 
 | |
1DG2
 
 | | SOLUTION CONFORMATION OF A-CONOTOXIN AUIB | | Descriptor: | A-CONOTOXIN AUIB | | Authors: | Cho, J.-H, Mok, K.H, Olivera, B.M, McIntosh, J.M, Park, K.-H, Han, K.-H. | | Deposit date: | 1999-11-23 | | Release date: | 2000-02-25 | | Last modified: | 2022-02-16 | | Method: | SOLUTION NMR | | Cite: | Nuclear magnetic resonance solution conformation of alpha-conotoxin AuIB, an alpha(3)beta(4) subtype-selective neuronal nicotinic acetylcholine receptor antagonist.
J.Biol.Chem., 275, 2000
|
|
4UV7
 
 | | The complex structure of extracellular domain of EGFR and GC1118A | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, EPIDERMAL GROWTH FACTOR RECEPTOR, ... | | Authors: | Yoo, J.H, Cho, H.S. | | Deposit date: | 2014-08-05 | | Release date: | 2015-10-14 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Gc1118, an Anti-Egfr Antibody with a Distinct Binding Epitope and Superior Inhibitory Activity Against High-Affinity Egfr Ligands.
Mol.Cancer Ther., 15, 2016
|
|
6J1U
 
 | | influenza virus nucleoprotein with a specific inhibitor | | Descriptor: | Nucleoprotein, ~{N}-[4-[(4-~{tert}-butylphenyl)carbonylamino]phenyl]-2,3-dihydro-1,4-benzodioxine-6-carboxamide | | Authors: | Pang, B, Zhang, W.Z, Zhang, H.M, Hao, Q. | | Deposit date: | 2018-12-29 | | Release date: | 2020-01-01 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Discovery of a Novel Specific Inhibitor Targeting Influenza A Virus Nucleoprotein with Pleiotropic Inhibitory Effects on Various Steps of the Viral Life Cycle.
J.Virol., 95, 2021
|
|
6GFX
 
 | | pVHL:EloB:EloC in complex with modified HIF-1a CODD peptide containing (3R,4S)-3-fluoro-4-hydroxyproline (ligand 13a) | | Descriptor: | Elongin-B, Elongin-C, FLUORINATED HYPOXIA-INDUCIBLE FACTOR 1 ALPHA PEPTIDE, ... | | Authors: | Castro, G.V, Testa, A, Ciulli, A. | | Deposit date: | 2018-05-02 | | Release date: | 2018-07-11 | | Last modified: | 2024-07-10 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | 3-Fluoro-4-hydroxyprolines: Synthesis, Conformational Analysis, and Stereoselective Recognition by the VHL E3 Ubiquitin Ligase for Targeted Protein Degradation.
J. Am. Chem. Soc., 140, 2018
|
|
6GFZ
 
 | | pVHL:EloB:EloC in complex with modified VH032 containing (3S,4S)-3-fluoro-4-hydroxyproline (ligand 14b) | | Descriptor: | (2~{R},3~{S},4~{S})-1-[(2~{S})-2-acetamido-3,3-dimethyl-butanoyl]-3-fluoranyl-~{N}-[[4-(4-methyl-1,3-thiazol-5-yl)phenyl]methyl]-4-oxidanyl-pyrrolidine-2-carboxamide, Elongin-B, Elongin-C, ... | | Authors: | Gadd, M.S, Testa, A, Ciulli, A. | | Deposit date: | 2018-05-02 | | Release date: | 2018-07-11 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | 3-Fluoro-4-hydroxyprolines: Synthesis, Conformational Analysis, and Stereoselective Recognition by the VHL E3 Ubiquitin Ligase for Targeted Protein Degradation.
J. Am. Chem. Soc., 140, 2018
|
|
