7O9T
 
 | |
7O9Z
 
 | | Crystal structure of Human Menin in complex with BD-08 | | Descriptor: | (~{E})-2-cyano-3-(2~{H}-indazol-6-yl)-~{N}-(2-morpholin-4-ylethyl)prop-2-enamide, 1,2-ETHANEDIOL, GLYCEROL, ... | | Authors: | Groves, M.R, Gao, K. | | Deposit date: | 2021-04-18 | | Release date: | 2022-07-27 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.979 Å) | | Cite: | Crystal structure of Human Menin in complex with BD-08
To Be Published
|
|
7O9X
 
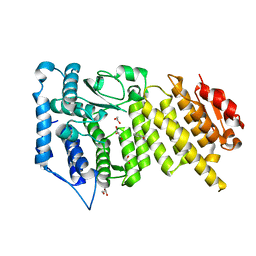 | |
7OA9
 
 | | Crystal structure of Human Menin in complex with Fragment 21 | | Descriptor: | (~{E})-2-cyano-3-(4-hydroxyphenyl)-~{N}-(2-morpholin-4-ylethyl)prop-2-enamide, 1,2-ETHANEDIOL, GLYCEROL, ... | | Authors: | Groves, M.R, Gao, K. | | Deposit date: | 2021-04-19 | | Release date: | 2022-08-03 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.098 Å) | | Cite: | Crystal structure of Human Menin in complex with Fragment 21
To Be Published
|
|
1PCI
 
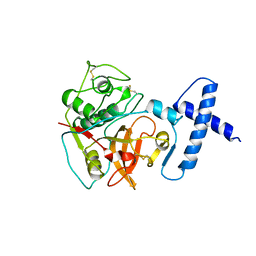 | | PROCARICAIN | | Descriptor: | PROCARICAIN | | Authors: | Groves, M.R, Taylor, M.A.J, Scott, M, Cummings, N.J, Pickersgill, R.W, Jenkins, J.A. | | Deposit date: | 1996-06-28 | | Release date: | 1997-04-01 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | The prosequence of procaricain forms an alpha-helical domain that prevents access to the substrate-binding cleft.
Structure, 4, 1996
|
|
2OB1
 
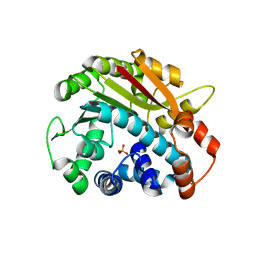 | | ppm1 with 1,8-ANS | | Descriptor: | Leucine carboxyl methyltransferase 1, PHOSPHATE ION | | Authors: | Groves, M.R. | | Deposit date: | 2006-12-18 | | Release date: | 2007-01-30 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | A method for the general identification of protein crystals in crystallization experiments using a noncovalent fluorescent dye.
Acta Crystallogr.,Sect.D, 63, 2007
|
|
2OB2
 
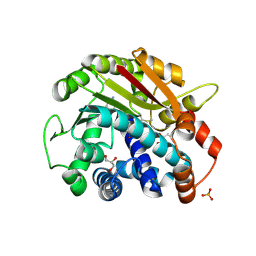 | | ppm1 in the absence of 1,8-ANS (cf 1JD) | | Descriptor: | GLYCEROL, Leucine carboxyl methyltransferase 1, PHOSPHATE ION, ... | | Authors: | Groves, M.R, Mueller, I.B, Kreplin, X, Mueller-Dieckmann, J. | | Deposit date: | 2006-12-18 | | Release date: | 2007-01-30 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | A method for the general identification of protein crystals in crystallization experiments using a noncovalent fluorescent dye.
Acta Crystallogr.,Sect.D, 63, 2007
|
|
1B3U
 
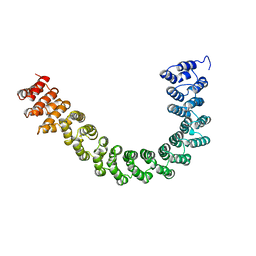 | | CRYSTAL STRUCTURE OF CONSTANT REGULATORY DOMAIN OF HUMAN PP2A, PR65ALPHA | | Descriptor: | PROTEIN (PROTEIN PHOSPHATASE PP2A) | | Authors: | Groves, M.R, Hanlon, N, Turowski, P, Hemmings, B, Barford, D. | | Deposit date: | 1998-12-14 | | Release date: | 1999-04-12 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The structure of the protein phosphatase 2A PR65/A subunit reveals the conformation of its 15 tandemly repeated HEAT motifs.
Cell(Cambridge,Mass.), 96, 1999
|
|
1CZI
 
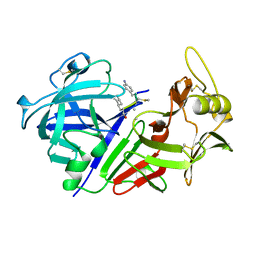 | | CHYMOSIN COMPLEX WITH THE INHIBITOR CP-113972 | | Descriptor: | CHYMOSIN, CP-113972 (NORSTATINE-S-METHYL CYSTEINE-IODO-PHENYLALANINE-PROLINE) | | Authors: | Groves, M.R, Dhanaraj, V, Pitts, J.E, Badasso, M, Hoover, D, Nugent, P, Blundell, T.L. | | Deposit date: | 1997-01-15 | | Release date: | 1997-04-01 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | A 2.3 A resolution structure of chymosin complexed with a reduced bond inhibitor shows that the active site beta-hairpin flap is rearranged when compared with the native crystal structure.
Protein Eng., 11, 1998
|
|
1BZC
 
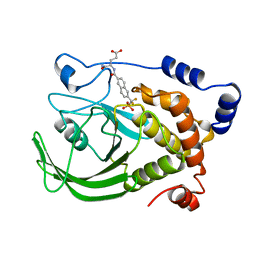 | | HUMAN PTP1B CATALYTIC DOMAIN COMPLEXED WITH TPI | | Descriptor: | 4-CARBAMOYL-4-{[6-(DIFLUORO-PHOSPHONO-METHYL)-NAPHTHALENE-2-CARBONYL]-AMINO}-BUTYRIC ACID, PROTEIN (PROTEIN-TYROSINE-PHOSPHATASE) | | Authors: | Groves, M.R, Yao, Z.J, Jr Burke, T.R, Barford, D. | | Deposit date: | 1998-10-27 | | Release date: | 1999-02-16 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structural basis for inhibition of the protein tyrosine phosphatase 1B by phosphotyrosine peptide mimetics.
Biochemistry, 37, 1998
|
|
1BZJ
 
 | | Human ptp1b complexed with tpicooh | | Descriptor: | 6-(DIFLUORO-PHOSPHONO-METHYL)-NAPHTHALENE-2-CARBOXYLIC ACID, PROTEIN (PROTEIN-TYROSINE-PHOSPHATASE) | | Authors: | Groves, M.R, Yao, Z.-J, Barford Jr, D.T.B. | | Deposit date: | 1998-10-29 | | Release date: | 1999-02-16 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structural basis for inhibition of the protein tyrosine phosphatase 1B by phosphotyrosine peptide mimetics.
Biochemistry, 37, 1998
|
|
1BZH
 
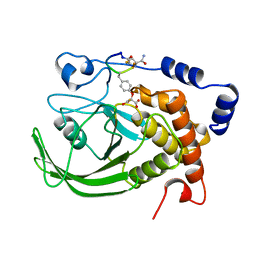 | | Cyclic peptide inhibitor of human PTP1B | | Descriptor: | PROTEIN (PROTEIN-TYROSINE-PHOSPHATASE 1B INHIBITOR), PROTEIN (PROTEIN-TYROSINE-PHOSPHATASE 1B) | | Authors: | Groves, M.R, Yao, Z.J, Burke Jr, T.R, Barford, D. | | Deposit date: | 1998-10-28 | | Release date: | 1999-02-16 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural basis for inhibition of the protein tyrosine phosphatase 1B by phosphotyrosine peptide mimetics.
Biochemistry, 37, 1998
|
|
6SKV
 
 | |
6S2K
 
 | | Human Menin in complex with AJ21 | | Descriptor: | Multiple endocrine neoplasia I, isoform CRA_b, ~{N}-[2-(4-methoxyphenyl)ethyl]-2-(4-nitrophenyl)imidazo[1,2-a]pyridin-3-amine | | Authors: | Groves, M.R, Gao, K. | | Deposit date: | 2019-06-21 | | Release date: | 2021-01-13 | | Last modified: | 2026-02-11 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Human Menin in complex with AJ21
To Be Published
|
|
6SKS
 
 | |
6SKT
 
 | |
3K7Y
 
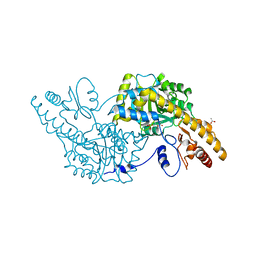 | | Aspartate Aminotransferase of Plasmodium falciparum | | Descriptor: | ACETATE ION, Aspartate aminotransferase, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Groves, M.R, Jordanova, R, Jain, R, Wrenger, C, Muller, I.B. | | Deposit date: | 2009-10-13 | | Release date: | 2010-10-13 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Specific Inhibition of the Aspartate Aminotransferase of Plasmodium falciparum.
J.Mol.Biol., 405, 2011
|
|
5NKZ
 
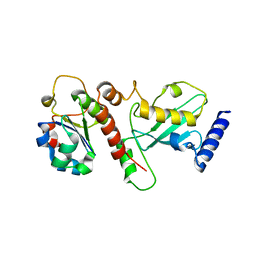 | | Crystal structure of H. polymorpha ubiquitin conjugating enzyme Pex4p in complex with soluble domain of Pex22p | | Descriptor: | Peroxin 22, Ubiquitin-conjugating enzyme E2-21 kDa | | Authors: | Danda, N, Lunev, S, Ali, A, Groves, M.R, Williams, C. | | Deposit date: | 2017-04-03 | | Release date: | 2018-01-24 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Structural insights into K48-linked ubiquitin chain formation by the Pex4p-Pex22p complex.
Biochem. Biophys. Res. Commun., 496, 2018
|
|
6SU9
 
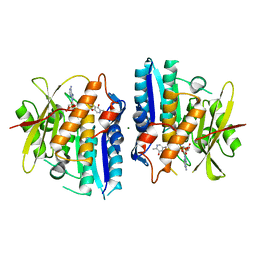 | |
6HL7
 
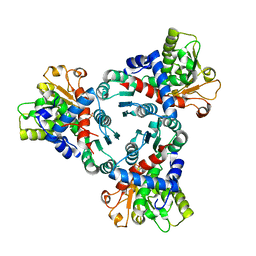 | | Crystal structure of truncated aspartate transcarbamoylase from Plasmodium falciparum with mutated active site (R109A/K138A) and N-carbamoyl-L-phosphate bound | | Descriptor: | Aspartate transcarbamoylase, PHOSPHORIC ACID MONO(FORMAMIDE)ESTER | | Authors: | Bosch, S.S, Lunev, S, Wrenger, C, Groves, M.R. | | Deposit date: | 2018-09-10 | | Release date: | 2018-09-26 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Molecular Target Validation of Aspartate Transcarbamoylase fromPlasmodium falciparumby Torin 2.
Acs Infect Dis., 6, 2020
|
|
8R2H
 
 | | Cryo-EM structure of 1-deoxy-D-xylulose 5-phosphate synthase (DXPS) from Plasmodium falciparum | | Descriptor: | 1-deoxy-D-xylulose-5-phosphate synthase, MAGNESIUM ION, THIAMINE DIPHOSPHATE | | Authors: | Gawriljuk, V.O, Godoy, A.S, Oerlemans, R, Groves, M.R. | | Deposit date: | 2023-11-06 | | Release date: | 2024-06-12 | | Last modified: | 2024-08-14 | | Method: | ELECTRON MICROSCOPY (2.42 Å) | | Cite: | Cryo-EM structure of 1-deoxy-D-xylulose 5-phosphate synthase DXPS from Plasmodium falciparum reveals a distinct N-terminal domain.
Nat Commun, 15, 2024
|
|
8Q5Z
 
 | |
2W9Y
 
 | | The structure of the lipid binding protein Ce-FAR-7 from Caenorhabditis elegans | | Descriptor: | FATTY ACID/RETINOL BINDING PROTEIN PROTEIN 7, ISOFORM A, CONFIRMED BY TRANSCRIPT EVIDENCE, ... | | Authors: | Jordanova, R, Groves, M.R, Tucker, P.A. | | Deposit date: | 2009-01-30 | | Release date: | 2009-10-20 | | Last modified: | 2025-04-09 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Fatty Acid and Retinoid Binding Proteins Have Distinct Binding Pockets for the Two Types of Cargo
J.Biol.Chem., 284, 2009
|
|
6FBA
 
 | | Crystal Structure of truncated aspartate transcarbamoylase from Plasmodium falciparum with bound inhibitor 2,3-naphthalenediol | | Descriptor: | Aspartate transcarbamoylase, DI(HYDROXYETHYL)ETHER, DIMETHYL SULFOXIDE, ... | | Authors: | Lunev, S, Bosch, S.S, Batista, F.A, Wang, C, Wrenger, C, Groves, M.R. | | Deposit date: | 2017-12-18 | | Release date: | 2018-02-21 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Identification of a non-competitive inhibitor of Plasmodium falciparum aspartate transcarbamoylase.
Biochem. Biophys. Res. Commun., 497, 2018
|
|
5MC7
 
 | |
