6WIW
 
 | | c-Src Bound to ATP-Competitive Inhibitor I14 | | Descriptor: | N-(3-{[4-amino-7-(2-methoxyethyl)-7H-pyrrolo[2,3-d]pyrimidin-5-yl]ethynyl}-4-methylphenyl)acetamide, Proto-oncogene tyrosine-protein kinase Src | | Authors: | Vilas-Boas, J, Thakur, M.K, Fang, L, Maly, D, Seeliger, M.A. | | Deposit date: | 2020-04-10 | | Release date: | 2020-06-17 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | How ATP-Competitive Inhibitors Allosterically Modulate Tyrosine Kinases That Contain a Src-like Regulatory Architecture.
Acs Chem.Biol., 15, 2020
|
|
4P08
 
 | | Engineered thermostable dimeric cocaine esterase | | Descriptor: | Cocaine esterase | | Authors: | Rodgers, D.W, Chow, K.-M, Fang, L, Zhan, C.-G. | | Deposit date: | 2014-02-20 | | Release date: | 2014-07-16 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.341 Å) | | Cite: | Rational design, preparation, and characterization of a therapeutic enzyme mutant with improved stability and function for cocaine detoxification.
Acs Chem.Biol., 9, 2014
|
|
3OXI
 
 | | Design and Synthesis of Disubstituted Thiophene and Thiazole Based Inhibitors of JNK for the Treatment of Neurodegenerative Diseases | | Descriptor: | Mitogen-activated protein kinase 10, Mitogen-activated protein kinase 8 interacting protein 1, methyl 3-[(thiophen-2-ylacetyl)amino]thiophene-2-carboxylate | | Authors: | Hom, R.K, Bowers, S, Sealy, J, Truong, A, Probst, G.D, Neitzel, M, Neitz, J, Fang, L, Brogley, L, Wu, J, Konradi, A.W, Sham, H, Toth, G, Pan, H, Yao, N, Artis, D.R. | | Deposit date: | 2010-09-21 | | Release date: | 2011-05-04 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Design and synthesis of disubstituted thiophene and thiazole based inhibitors of JNK.
Bioorg.Med.Chem.Lett., 20, 2010
|
|
5ER4
 
 | | Crystal Structure of Calcium-loaded S100B bound to SC0025 | | Descriptor: | 6-methyl-5,6,6~{a},7-tetrahydro-4~{H}-dibenzo[de,g]quinoline-10,11-diol, CALCIUM ION, Protein S100-B | | Authors: | Cavalier, M.C, Melville, Z.E, Aligholizadeh, E, Fang, L, Alasady, M.J, Pierce, A.D, Wilder, P.T, MacKerell Jr, A.D, Weber, D.J. | | Deposit date: | 2015-11-13 | | Release date: | 2016-06-08 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.813 Å) | | Cite: | Novel protein-inhibitor interactions in site 3 of Ca(2+)-bound S100B as discovered by X-ray crystallography.
Acta Crystallogr D Struct Biol, 72, 2016
|
|
5ER5
 
 | | Crystal Structure of Calcium-loaded S100B bound to SC1990 | | Descriptor: | CALCIUM ION, ETHIDIUM, Protein S100-B | | Authors: | Cavalier, M.C, Melville, Z.E, Aligholizadeh, E, Fang, L, Alasady, M.J, Pierce, A.D, Wilder, P.T, MacKerell Jr, A.D, Weber, D.J. | | Deposit date: | 2015-11-13 | | Release date: | 2016-06-08 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.26 Å) | | Cite: | Novel protein-inhibitor interactions in site 3 of Ca(2+)-bound S100B as discovered by X-ray crystallography.
Acta Crystallogr D Struct Biol, 72, 2016
|
|
5Y4L
 
 | | PRRSV nsp4 | | Descriptor: | Non-structural protein | | Authors: | Shi, Y.J, Gang, Y, Peng, G.Q. | | Deposit date: | 2017-08-04 | | Release date: | 2018-01-17 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.198 Å) | | Cite: | Identification of two antiviral inhibitors targeting 3C-like serine/3C-like protease of porcine reproductive and respiratory syndrome virus and porcine epidemic diarrhea virus.
Vet. Microbiol., 213, 2018
|
|
6T8U
 
 | | Complement factor B in complex with 5-Bromo-3-chloro-N-(4,5-dihydro-1H-imidazol-2-yl)-7-methyl-1H-indol-4-amine | | Descriptor: | 5-bromanyl-3-chloranyl-~{N}-(1~{H}-imidazol-2-yl)-7-methyl-1~{H}-indol-4-amine, Complement factor B, SULFATE ION | | Authors: | Mainolfi, N, Ehara, T, Karki, R.G, Anderson, K, Mac Sweeney, A, Wiesmann, C, Adams, C, Liao, S.-M, Argikar, U.A, Jendza, K, Zhang, C, Powers, J, Klosowski, D.W, Crowley, M, Kawanami, T, Ding, J, April, M, Forster, C, Serrano-Wu, M, Capparelli, M, Ramqaj, R, Solovay, C, Cumin, F, Smith, T.M, Ferrara, L, Lee, W, Long, D, Prentiss, M, De Erkenez, A, Yang, L, Fang, L, Sellner, H, Sirockin, F, Valeur, E, Erbel, P, Ramage, P, Gerhartz, B, Schubart, A, Flohr, S, Gradoux, N, Feifel, R, Vogg, B, Maibaum, J, Eder, J, Sedrani, R, Harrison, R.A, Mogi, M, Jaffee, B.D, Adams, C.M. | | Deposit date: | 2019-10-25 | | Release date: | 2020-03-04 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.84 Å) | | Cite: | Discovery of 4-((2S,4S)-4-Ethoxy-1-((5-methoxy-7-methyl-1H-indol-4-yl)methyl)piperidin-2-yl)benzoic Acid (LNP023), a Factor B Inhibitor Specifically Designed To Be Applicable to Treating a Diverse Array of Complement Mediated Diseases.
J.Med.Chem., 63, 2020
|
|
6T8V
 
 | | Complement factor B in complex with (S)-5,7-Dimethyl-4-((2-phenylpiperidin-1-yl)methyl)-1H-indole | | Descriptor: | 4-[(2~{S})-1-[(5,7-dimethyl-1~{H}-indol-4-yl)methyl]piperidin-2-yl]benzoic acid, Complement factor B, SULFATE ION, ... | | Authors: | Mainolfi, N, Ehara, T, Karki, R.G, Anderson, K, Mac Sweeney, A, Wiesmann, C, Adams, C, Mainolfi, N, Liao, S.-M, Argikar, U.A, Jendza, K, Zhang, C, Powers, J, Klosowski, D.W, Crowley, M, Kawanami, T, Ding, J, April, M, Forster, C, Serrano-Wu, M, Capparelli, M, Ramqaj, R, Solovay, C, Cumin, F, Smith, T.M, Ferrara, L, Lee, W, Long, D, Prentiss, M, De Erkenez, A, Yang, L, Fang, L, Sellner, H, Sirockin, F, Valeur, E, Erbel, P, Ramage, P, Gerhartz, B, Schubart, A, Flohr, S, Gradoux, N, Feifel, R, Vogg, B, Wiesmann, C, Maibaum, J, Eder, J, Sedrani, R, Harrison, R.A, Mogi, M, Jaffee, B.D, Adams, C.M. | | Deposit date: | 2019-10-25 | | Release date: | 2020-03-04 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | Discovery of 4-((2S,4S)-4-Ethoxy-1-((5-methoxy-7-methyl-1H-indol-4-yl)methyl)piperidin-2-yl)benzoic Acid (LNP023), a Factor B Inhibitor Specifically Designed To Be Applicable to Treating a Diverse Array of Complement Mediated Diseases.
J.Med.Chem., 63, 2020
|
|
7OXL
 
 | | Crystal structure of human Spermine Oxidase | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CHLORIDE ION, FAD-MDL72527 adduct, ... | | Authors: | Impagliazzo, A, Johannsson, S, Thomsen, M, Krapp, S. | | Deposit date: | 2021-06-22 | | Release date: | 2022-07-13 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure of human spermine oxidase in complex with a highly selective allosteric inhibitor.
Commun Biol, 5, 2022
|
|
7OY0
 
 | | Structure of human Spermine Oxidase in complex with a highly selective allosteric inhibitor | | Descriptor: | 4-[(4-imidazo[1,2-a]pyridin-3-yl-1,3-thiazol-2-yl)amino]phenol, CHLORIDE ION, FAD-MDL72527 adduct, ... | | Authors: | Impagliazzo, A, Thomsen, M, Johannsson, S, Krapp, S. | | Deposit date: | 2021-06-23 | | Release date: | 2022-07-13 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Structure of human spermine oxidase in complex with a highly selective allosteric inhibitor.
Commun Biol, 5, 2022
|
|
6T8W
 
 | | Complement factor B in complex with (-)-4-(1-((5,7-Dimethyl-1H-indol-4-yl)methyl)piperidin-2-yl)benzoic acid | | Descriptor: | 5,7-dimethyl-4-[[(2~{S})-2-phenylpiperidin-1-yl]methyl]-1~{H}-indole, Complement factor B, SULFATE ION, ... | | Authors: | Mainolfi, N, Ehara, T, Karki, R.G, Anderson, K, Sweeney, A.M, Wiesmann, C, Adams, C, Mainolfi, N, Liao, S.M, Argikar, U.A, Jendza, K, Zhang, C, Powers, J, Klosowski, D.W, Crowley, M, Kawanami, T, Ding, J, April, M, Forster, C, Wu, M.S, Capparelli, M, Ramqaj, R, Solovay, C, Cumin, F, Smith, T.M, Ferrara, L, Lee, W, Long, D, Prentiss, M, Erkenez, A.D, Yang, L, Fang, L, Sellner, H, Sirockin, F, Valeur, E, Erbel, P, Ramage, P, Gerhartz, B, Schubart, A, Flohr, S, Gradoux, N, Feifel, R, Vogg, B, Wiesmann, C, Maibaum, J, Eder, J, Sedrani, R, Harrison, R.A, Mogi, M, Jaffee, B.D, Adams, C.M. | | Deposit date: | 2019-10-25 | | Release date: | 2020-03-04 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Discovery of 4-((2S,4S)-4-Ethoxy-1-((5-methoxy-7-methyl-1H-indol-4-yl)methyl)piperidin-2-yl)benzoic Acid (LNP023), a Factor B Inhibitor Specifically Designed To Be Applicable to Treating a Diverse Array of Complement Mediated Diseases.
J.Med.Chem., 63, 2020
|
|
5NNW
 
 | | NLPPya in complex with glucosamine | | Descriptor: | 2-amino-2-deoxy-beta-D-glucopyranose, 25 kDa protein elicitor, MAGNESIUM ION | | Authors: | Podobnik, M, Anderluh, G, Lenarcic, T. | | Deposit date: | 2017-04-10 | | Release date: | 2017-12-27 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Eudicot plant-specific sphingolipids determine host selectivity of microbial NLP cytolysins.
Science, 358, 2017
|
|
5NO9
 
 | | NLPPya in complex with mannosamine | | Descriptor: | 2-amino-2-deoxy-alpha-D-mannopyranose, 25 kDa protein elicitor, MAGNESIUM ION | | Authors: | Podobnik, M, Anderluh, G, Lenarcic, T. | | Deposit date: | 2017-04-11 | | Release date: | 2017-12-27 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Eudicot plant-specific sphingolipids determine host selectivity of microbial NLP cytolysins.
Science, 358, 2017
|
|
8D4Z
 
 | | Crystal structure of USP7 in complex with allosteric inhibitor FX1-3763 | | Descriptor: | 1-({(7M)-7-[1-(azetidin-3-yl)-6-chloro-1,2,3,4-tetrahydroquinolin-8-yl]thieno[3,2-b]pyridin-2-yl}methyl)pyrrolidine-2,5-dione, GLYCEROL, Ubiquitin carboxyl-terminal hydrolase 7 | | Authors: | Bell, J.A. | | Deposit date: | 2022-06-03 | | Release date: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | Novel USP7 inhibitors demonstrate potent anti-cancer activity in models of AML, synergy with BCL2 inhibition, and a differentiated mechanism of action
To Be Published
|
|
6WG9
 
 | | Crystal structure of tetracycline destructase Tet(X7) | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Tetracycline destructase Tet(X7) | | Authors: | Kumar, H, Tolia, N.H. | | Deposit date: | 2020-04-05 | | Release date: | 2021-02-10 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Tetracycline-inactivating enzymes from environmental, human commensal, and pathogenic bacteria cause broad-spectrum tetracycline resistance.
Commun Biol, 3, 2020
|
|
6ZBQ
 
 | | Small-molecule inhibitors of the PDZ domain of Dishevelled proteins interrupt Wnt signalling | | Descriptor: | 5-fluoranyl-2-(5,6,7,8-tetrahydronaphthalen-2-ylsulfonylamino)benzoic acid, Dishevelled, dsh homolog 3 (Drosophila), ... | | Authors: | Roske, Y, Heinemann, U, Oschkinat, H. | | Deposit date: | 2020-06-09 | | Release date: | 2021-06-23 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.43 Å) | | Cite: | Small-molecule inhibitors of the PDZ domain of Dishevelled proteins interrupt Wnt signalling
J.Magn.Reson., 2021
|
|
6ZBZ
 
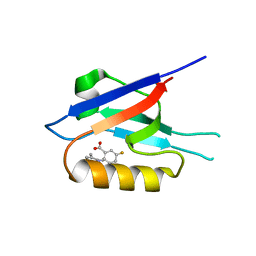 | | Small-molecule inhibitors of the PDZ domain of Dishevelled proteins interrupt Wnt signalling | | Descriptor: | 1,2-ETHANEDIOL, 2-[(3,4-dimethylphenyl)sulfonylamino]-5-fluoranyl-benzoic acid, Dishevelled, ... | | Authors: | Roske, Y, Heinemann, U, Oschkinat, H. | | Deposit date: | 2020-06-09 | | Release date: | 2021-06-23 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Small-molecule inhibitors of the PDZ domain of Dishevelled proteins interrupt Wnt signalling
J.Magn.Reson., 2021
|
|
6ZC7
 
 | |
6ZC6
 
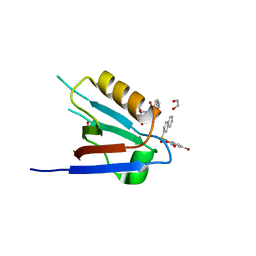 | |
6ZC3
 
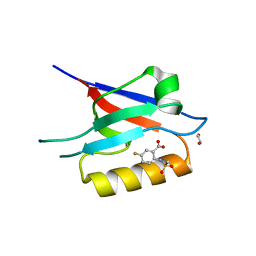 | | Small-molecule inhibitors of the PDZ domain of Dishevelled proteins interrupt Wnt signalling | | Descriptor: | 1,2-ETHANEDIOL, 5-fluoranyl-2-(methylsulfonylamino)benzoic acid, Dishevelled, ... | | Authors: | Roske, Y, Heinemann, U, Oschkinat, H. | | Deposit date: | 2020-06-09 | | Release date: | 2021-06-23 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Small-molecule inhibitors of the PDZ domain of Dishevelled proteins interrupt Wnt signalling
J.Magn.Reson., 2021
|
|
6ZC8
 
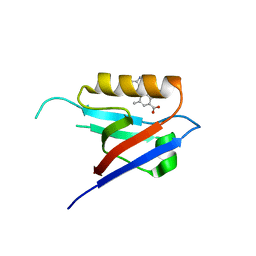 | |
6ZC4
 
 | | Small-molecule inhibitors of the PDZ domain of Dishevelled proteins interrupt Wnt signalling | | Descriptor: | 5-bromanyl-2-(5,6,7,8-tetrahydronaphthalen-2-ylsulfonylamino)benzoic acid, Dishevelled, dsh homolog 3 (Drosophila), ... | | Authors: | Roske, Y, Heinemann, U, Oschkinat, H. | | Deposit date: | 2020-06-09 | | Release date: | 2021-06-23 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Small-molecule inhibitors of the PDZ domain of Dishevelled proteins interrupt Wnt signalling
J.Magn.Reson., 2021
|
|
3N4L
 
 | | BACE-1 in complex with ELN380842 | | Descriptor: | Beta-secretase 1, N-[(1S,2R)-1-(3,5-difluorobenzyl)-2-hydroxy-3-({1-[3-(1H-pyrazol-1-yl)phenyl]cyclohexyl}amino)propyl]acetamide | | Authors: | Yao, N.H. | | Deposit date: | 2010-05-21 | | Release date: | 2010-11-24 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Improving the permeability of the hydroxyethylamine BACE-1 inhibitors: structure-activity relationship of P2' substituents.
Bioorg.Med.Chem.Lett., 20, 2010
|
|
5HIZ
 
 | | The structure of PEDV NSP9 | | Descriptor: | Non-structural protein 9 | | Authors: | Deng, F, Peng, G. | | Deposit date: | 2016-01-12 | | Release date: | 2017-01-25 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Dimerization of Coronavirus nsp9 with Diverse Modes Enhances Its Nucleic Acid Binding Affinity.
J.Virol., 92, 2018
|
|
5HIY
 
 | | Crystal structure of PEDV NSP9 Mutant-C59A | | Descriptor: | Non-structural protein 9 | | Authors: | Deng, F, Peng, G. | | Deposit date: | 2016-01-12 | | Release date: | 2017-01-25 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Dimerization of Coronavirus nsp9 with Diverse Modes Enhances Its Nucleic Acid Binding Affinity.
J.Virol., 92, 2018
|
|
