4LGE
 
 | | Crystal structure of clAP1 BIR3 bound to T3261256 | | Descriptor: | (4S,6aR,7aS)-5-{(2S)-2-cyclohexyl-2-[(N-methyl-L-alanyl)amino]acetyl}-N-[(4R)-3,4-dihydro-2H-chromen-4-yl]octahydro-1H-cyclopropa[4,5]pyrrolo[1,2-a]pyrazine-4-carboxamide, Baculoviral IAP repeat-containing protein 2, ZINC ION | | Authors: | Dougan, D.R. | | Deposit date: | 2013-06-27 | | Release date: | 2013-08-28 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Design, stereoselective synthesis, and biological evaluation of novel tri-cyclic compounds as inhibitor of apoptosis proteins (IAP) antagonists.
Bioorg.Med.Chem., 21, 2013
|
|
4LGU
 
 | | Crystal structure of clAP1 BIR3 bound to T3226692 | | Descriptor: | (3S,8aR)-N-((R)-chroman-4-yl)-2-((S)-2-cyclohexyl-2-((S)-2-(methylamino)propanamido)acetyl)octahydropyrrolo[1,2-a]pyrazine-3-carboxamide, Baculoviral IAP repeat-containing protein 2, ZINC ION | | Authors: | Dougan, D.R, Mol, C.D, Snell, G.P. | | Deposit date: | 2013-06-28 | | Release date: | 2013-08-28 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Design, stereoselective synthesis, and biological evaluation of novel tri-cyclic compounds as inhibitor of apoptosis proteins (IAP) antagonists.
Bioorg.Med.Chem., 21, 2013
|
|
8VQX
 
 | |
8GMN
 
 | | Crystal structure of human C1s in complex with inhibitor | | Descriptor: | Complement C1s subcomponent, SULFATE ION, [4-(1-aminophthalazin-6-yl)piperazin-1-yl](2-methylphenyl)methanone | | Authors: | Dougan, D.R, Lane, W. | | Deposit date: | 2023-03-26 | | Release date: | 2023-05-17 | | Last modified: | 2023-05-24 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Discovery of a Novel Series of Potent, Selective, Orally Available, and Brain-Penetrable C1s Inhibitors for Modulation of the Complement Pathway.
J.Med.Chem., 66, 2023
|
|
3MBL
 
 | |
5JFR
 
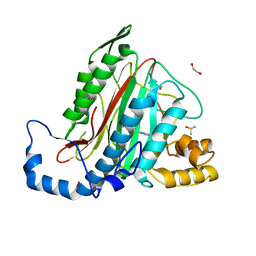 | | Potent, Reversible MetAP2 Inhibitors via Fragment Based Drug Discovery | | Descriptor: | 1,2-ETHANEDIOL, 7-fluoro-4-(5-methyl-3H-imidazo[4,5-b]pyridin-6-yl)-2,4-dihydropyrazolo[4,3-b]indole, DIMETHYL SULFOXIDE, ... | | Authors: | Dougan, D.R, Lawson, J.D. | | Deposit date: | 2016-04-19 | | Release date: | 2016-05-25 | | Last modified: | 2016-06-15 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Discovery of potent, reversible MetAP2 inhibitors via fragment based drug discovery and structure based drug design-Part 2.
Bioorg.Med.Chem.Lett., 26, 2016
|
|
3TDC
 
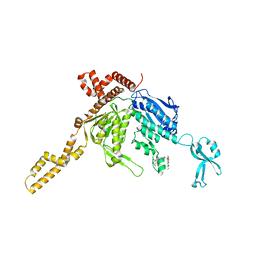 | | Crystal Structure of Human Acetyl-CoA carboxylase 2 | | Descriptor: | 1-[3-({4-[(5S)-3,3-dimethyl-1-oxo-2-oxa-7-azaspiro[4.5]dec-7-yl]piperidin-1-yl}carbonyl)-1-benzothiophen-2-yl]-3-ethylurea, Acetyl-CoA carboxylase 2 variant | | Authors: | Dougan, D.R, Mol, C.D. | | Deposit date: | 2011-08-10 | | Release date: | 2011-10-12 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.41 Å) | | Cite: | Design, synthesis, and structure-activity relationships of spirolactones bearing 2-ureidobenzothiophene as acetyl-CoA carboxylases inhibitors.
Bioorg.Med.Chem.Lett., 21, 2011
|
|
5JHU
 
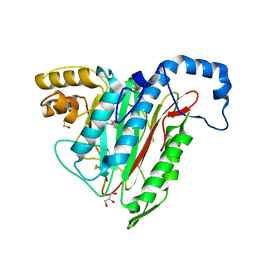 | | Potent, Reversible MetAP2 Inhibitors via FBDD | | Descriptor: | DIMETHYL SULFOXIDE, GLYCEROL, MANGANESE (II) ION, ... | | Authors: | Dougan, D.R. | | Deposit date: | 2016-04-21 | | Release date: | 2016-05-25 | | Last modified: | 2017-11-22 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Discovery of potent, reversible MetAP2 inhibitors via fragment based drug discovery and structure based drug design-Part 1.
Bioorg.Med.Chem.Lett., 26, 2016
|
|
5JI6
 
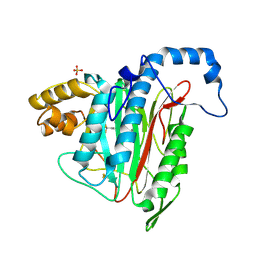 | | Potent, Reversible MetAP2 Inhibitors via FBDD | | Descriptor: | 4-(3-methylpyridin-4-yl)-6-(trifluoromethyl)-1H-indazole, MANGANESE (II) ION, Methionine aminopeptidase 2, ... | | Authors: | Dougan, D.R, Lawson, J.D. | | Deposit date: | 2016-04-21 | | Release date: | 2016-05-25 | | Last modified: | 2017-11-22 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Discovery of potent, reversible MetAP2 inhibitors via fragment based drug discovery and structure based drug design-Part 1.
Bioorg.Med.Chem.Lett., 26, 2016
|
|
4Z3V
 
 | |
4ZLY
 
 | |
4ZLZ
 
 | |
6XIH
 
 | |
4HY5
 
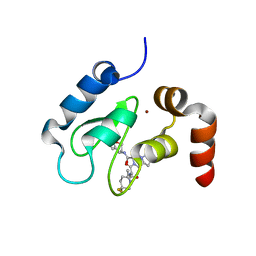 | | Crystal structure of cIAP1 BIR3 bound to T3256336 | | Descriptor: | (3S,7R,8aR)-2-{(2S)-2-(4,4-difluorocyclohexyl)-2-[(N-methyl-L-alanyl)amino]acetyl}-N-[(4R)-3,4-dihydro-2H-chromen-4-yl]-7-ethoxyoctahydropyrrolo[1,2-a]pyrazine-3-carboxamide, Baculoviral IAP repeat-containing protein 2, ZINC ION | | Authors: | Dougan, D.R, Snell, G.P. | | Deposit date: | 2012-11-13 | | Release date: | 2013-01-30 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.752 Å) | | Cite: | Design and Synthesis of Potent Inhibitor of Apoptosis (IAP) Proteins Antagonists Bearing an Octahydropyrrolo[1,2-a]pyrazine Scaffold as a Novel Proline Mimetic.
J.Med.Chem., 56, 2013
|
|
4HY4
 
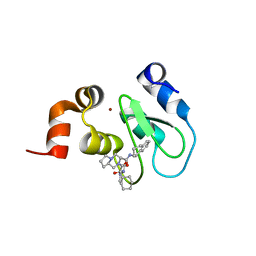 | | Crystal structure of cIAP1 BIR3 bound to T3170284 | | Descriptor: | (3S,8aR)-2-{(2S)-2-cyclohexyl-2-[(N-methyl-L-alanyl)amino]acetyl}-N-[(1R)-1,2,3,4-tetrahydronaphthalen-1-yl]octahydropyrrolo[1,2-a]pyrazine-3-carboxamide, Baculoviral IAP repeat-containing protein 2, ZINC ION | | Authors: | Dougan, D.R, Mol, C.D, Snell, G.P. | | Deposit date: | 2012-11-12 | | Release date: | 2013-01-30 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.249 Å) | | Cite: | Design and Synthesis of Potent Inhibitor of Apoptosis (IAP) Proteins Antagonists Bearing an Octahydropyrrolo[1,2-a]pyrazine Scaffold as a Novel Proline Mimetic.
J.Med.Chem., 56, 2013
|
|
4L6S
 
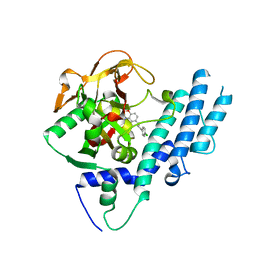 | | PARP complexed with benzo[1,4]oxazin-3-one inhibitor | | Descriptor: | (2S)-6-{[4-(4-chlorophenyl)-3,6-dihydropyridin-1(2H)-yl]methyl}-2-methyl-2H-1,4-benzoxazin-3(4H)-one, Poly [ADP-ribose] polymerase 1 | | Authors: | Dougan, D.R, Mol, C.D, Lawson, J.D. | | Deposit date: | 2013-06-12 | | Release date: | 2013-08-07 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Discovery of novel benzo[b][1,4]oxazin-3(4H)-ones as poly(ADP-ribose)polymerase inhibitors
Bioorg.Med.Chem.Lett., 23, 2013
|
|
3PP1
 
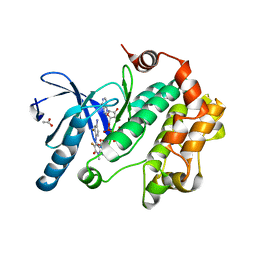 | | Crystal Structure of the Human Mitogen-activated protein kinase kinase 1 (MEK 1) in complex with ligand and MgATP | | Descriptor: | 3-[(2R)-2,3-dihydroxypropyl]-6-fluoro-5-[(2-fluoro-4-iodophenyl)amino]-8-methylpyrido[2,3-d]pyrimidine-4,7(3H,8H)-dione, ACETATE ION, ADENOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Dougan, D.R. | | Deposit date: | 2010-11-23 | | Release date: | 2011-02-23 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Discovery of TAK-733, a potent and selective MEK allosteric site inhibitor for the treatment of cancer.
Bioorg.Med.Chem.Lett., 21, 2011
|
|
5V24
 
 | |
5UOX
 
 | |
5USQ
 
 | | ALK-5 kinase inhibitor complex | | Descriptor: | N-[2-(5-chloro-2-fluorophenyl)pyridin-4-yl]-2-[(piperidin-4-yl)methyl]-2H-pyrazolo[4,3-b]pyridin-7-amine, TGF-beta receptor type-1 | | Authors: | Dougan, D.R, Lawson, J.D. | | Deposit date: | 2017-02-13 | | Release date: | 2017-04-12 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Design, synthesis and optimization of 7-substituted-pyrazolo[4,3-b]pyridine ALK5 (activin receptor-like kinase 5) inhibitors.
Bioorg. Med. Chem. Lett., 27, 2017
|
|
5UOR
 
 | | Structure-Based Design of ASK1 Inhibitors as Potential First-in-Class Agents for Heart Failure | | Descriptor: | 6-{[(2S)-4-methylmorpholin-2-yl]methoxy}-2-{6-[4-(propan-2-yl)-4H-1,2,4-triazol-3-yl]pyridin-2-yl}-2,3-dihydro-1H-isoindol-1-one, Mitogen-activated protein kinase kinase kinase 5, SULFATE ION | | Authors: | Dougan, D.R. | | Deposit date: | 2017-02-01 | | Release date: | 2017-06-07 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Structure-Based Design of ASK1 Inhibitors as Potential Agents for Heart Failure.
ACS Med Chem Lett, 8, 2017
|
|
5W49
 
 | | The crystal structure of human S-adenosylhomocysteine hydrolase (AHCY) bound to oxadiazole inhibitor | | Descriptor: | (4-amino-1,2,5-oxadiazol-3-yl)[(3R)-3-{4-[(3-methoxyphenyl)amino]-6-methylpyridin-2-yl}pyrrolidin-1-yl]methanone, 1,2-ETHANEDIOL, Adenosylhomocysteinase, ... | | Authors: | Dougan, D.R, Lawson, J.D, Lane, W. | | Deposit date: | 2017-06-09 | | Release date: | 2017-06-28 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Identification of AHCY inhibitors using novel high-throughput mass spectrometry.
Biochem. Biophys. Res. Commun., 491, 2017
|
|
5UP3
 
 | |
7N5O
 
 | | Fragment-Based Discovery of a Novel Bruton's Tyrosine Kinase Inhibitor | | Descriptor: | 1,2-ETHANEDIOL, 5-(1H-benzimidazol-2-yl)-2,4-dihydro-3H-1,2,4-triazol-3-one, IMIDAZOLE, ... | | Authors: | Dougan, D.R, Lawson, J.D. | | Deposit date: | 2021-06-06 | | Release date: | 2021-09-08 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Discovery of the Bruton's Tyrosine Kinase Inhibitor Clinical Candidate TAK-020 ( S )-5-(1-((1-Acryloylpyrrolidin-3-yl)oxy)isoquinolin-3-yl)-2,4-dihydro-3 H -1,2,4-triazol-3-one, by Fragment-Based Drug Design.
J.Med.Chem., 64, 2021
|
|
7N5R
 
 | | Fragment-Based Discovery of a Novel Bruton's Tyrosine Kinase Inhibitor | | Descriptor: | 1,2-ETHANEDIOL, 5-phenyl-2,4-dihydro-3H-1,2,4-triazol-3-one, DIMETHYL SULFOXIDE, ... | | Authors: | Dougan, D.R, Lawson, J.D. | | Deposit date: | 2021-06-06 | | Release date: | 2021-09-08 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Discovery of the Bruton's Tyrosine Kinase Inhibitor Clinical Candidate TAK-020 ( S )-5-(1-((1-Acryloylpyrrolidin-3-yl)oxy)isoquinolin-3-yl)-2,4-dihydro-3 H -1,2,4-triazol-3-one, by Fragment-Based Drug Design.
J.Med.Chem., 64, 2021
|
|
