1H1R
 
 | | Structure of human Thr160-phospho CDK2/cyclin A complexed with the inhibitor NU6086 | | Descriptor: | 6-CYCLOHEXYLMETHOXY-2-(3'-CHLOROANILINO) PURINE, CELL DIVISION PROTEIN KINASE 2, CYCLIN A2 | | Authors: | Davies, T.G, Noble, M.E.M, Endicott, J.A, Johnson, L.N. | | Deposit date: | 2002-07-21 | | Release date: | 2002-09-19 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure-based design of a potent purine-based cyclin-dependent kinase inhibitor.
Nat. Struct. Biol., 9, 2002
|
|
1H1Q
 
 | | Structure of human Thr160-phospho CDK2/cyclin A complexed with the inhibitor NU6094 | | Descriptor: | 2-ANILINO-6-CYCLOHEXYLMETHOXYPURINE, CELL DIVISION PROTEIN KINASE 2, CYCLIN A2 | | Authors: | Davies, T.G, Noble, M.E.M, Endicott, J.A, Johnson, L.N. | | Deposit date: | 2002-07-21 | | Release date: | 2002-09-19 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure-Based Design of a Potent Purine-Based Cyclin-Dependent Kinase Inhibitor
Nat.Struct.Biol., 9, 2002
|
|
1H1P
 
 | | Structure of human Thr160-phospho CDK2/cyclin A complexed with the inhibitor NU2058 | | Descriptor: | 6-O-CYCLOHEXYLMETHYL GUANINE, CELL DIVISION PROTEIN KINASE 2, CYCLIN A2 | | Authors: | Davies, T.G, Noble, M.E.M, Endicott, J.A, Johnson, L.N. | | Deposit date: | 2002-07-21 | | Release date: | 2002-09-19 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure-Based Design of a Potent Purine-Based Cyclin-Dependent Kinase Inhibitor
Nat.Struct.Biol., 9, 2002
|
|
1H1S
 
 | | Structure of human Thr160-phospho CDK2/cyclin A complexed with the inhibitor NU6102 | | Descriptor: | CELL DIVISION PROTEIN KINASE 2, CYCLIN A2, O6-CYCLOHEXYLMETHOXY-2-(4'-SULPHAMOYLANILINO) PURINE | | Authors: | Davies, T.G, Noble, M.E.M, Endicott, J.A, Johnson, L.N. | | Deposit date: | 2002-07-21 | | Release date: | 2002-09-19 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure-Based Design of a Potent Purine-Based Cyclin-Dependent Kinase Inhibitor
Nat.Struct.Biol., 9, 2002
|
|
2JDS
 
 | | Structure of cAMP-dependent protein kinase complexed with A-443654 | | Descriptor: | (2S)-1-(1H-INDOL-3-YL)-3-{[5-(3-METHYL-1H-INDAZOL-5-YL)PYRIDIN-3-YL]OXY}PROPAN-2-AMINE, CAMP-DEPENDENT PROTEIN KINASE, CAMP-DEPENDENT PROTEIN KINASE INHIBITOR ALPHA | | Authors: | Davies, T.G, Verdonk, M.L, Graham, B, Saalau-Bethell, S, Hamlett, C.C.F, McHardy, T, Collins, I, Garrett, M.D, Workman, P, Woodhead, S.J, Jhoti, H, Barford, D. | | Deposit date: | 2007-01-12 | | Release date: | 2007-02-13 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A Structural Comparison of Inhibitor Binding to Pkb, Pka and Pka-Pkb Chimera
J.Mol.Biol., 367, 2007
|
|
2JDT
 
 | | Structure of PKA-PKB chimera complexed with ISOQUINOLINE-5-SULFONIC ACID (2-(2-(4-CHLOROBENZYLOXY) ETHYLAMINO)ETHYL)AMIDE | | Descriptor: | CAMP-DEPENDENT PROTEIN KINASE, CAMP-DEPENDENT PROTEIN KINASE INHIBITOR ALPHA, ISOQUINOLINE-5-SULFONIC ACID (2-(2-(4-CHLOROBENZYLOXY)ETHYLAMINO)ETHYL)AMIDE | | Authors: | Davies, T.G, Verdonk, M.L, Graham, B, Saalau-Bethell, S, Hamlett, C.C.F, McHardy, T, Collins, I, Garrett, M.D, Workman, P, Woodhead, S.J, Jhoti, H, Barford, D. | | Deposit date: | 2007-01-12 | | Release date: | 2007-02-13 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | A Structural Comparison of Inhibitor Binding to Pkb, Pka and Pka-Pkb Chimera
J.Mol.Biol., 367, 2007
|
|
5FNQ
 
 | | Structure of the Keap1 Kelch domain in complex with a small molecule inhibitor. | | Descriptor: | 3-(4-CHLOROPHENYL)PROPANOIC ACID, KELCH-LIKE ECH-ASSOCIATED PROTEIN 1 | | Authors: | Davies, T.G, Wixted, W.E, Coyle, J.E, Griffiths-Jones, C, Hearn, K, McMenamin, R, Norton, D, Rich, S.J, Richardson, C, Saxty, G, Willems, H.M.G, Woolford, A.J, Cottom, J.E, Kou, J, Yonchuk, J.G, Feldser, H.G, Sanchez, Y, Foley, J.P, Bolognese, B.J, Logan, G, Podolin, P.L, Yan, H, Callahan, J.F, Heightman, T.D, Kerns, J.K. | | Deposit date: | 2015-11-16 | | Release date: | 2016-04-13 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Mono-Acidic Inhibitors of the Kelch-Like Ech-Associated Protein 1 : Nuclear Factor Erythroid 2-Related Factor 2 (Keap1:Nrf2) Protein-Protein Interaction with High Cell Potency Identified by Fragment-Based Discovery.
J.Med.Chem., 59, 2016
|
|
5FZN
 
 | | Structure of the Keap1 Kelch domain in complex with a small molecule inhibitor. | | Descriptor: | KELCH-LIKE ECH-ASSOCIATED PROTEIN 1, SULFATE ION, benzenesulfonamide | | Authors: | Davies, T.G, Wixted, W.E, Coyle, J.E, Griffiths-Jones, C, Hearn, K, McMenamin, R, Norton, D, Rich, S.J, Richardson, C, Saxty, G, Willems, H.M.G, Woolford, A.J, Cottom, J.E, Kou, J, Yonchuk, J.G, Feldser, H.G, Sanchez, Y, Foley, J.P, Bolognese, B.J, Logan, G, Podolin, P.L, Yan, H, Callahan, J.F, Heightman, T.D, Kerns, J.K. | | Deposit date: | 2016-03-15 | | Release date: | 2016-04-13 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Mono-Acidic Inhibitors of the Kelch-Like Ech-Associated Protein 1 : Nuclear Factor Erythroid 2-Related Factor 2 (Keap1:Nrf2) Protein-Protein Interaction with High Cell Potency Identified by Fragment-Based Discovery.
J.Med.Chem., 59, 2016
|
|
2JDO
 
 | | STRUCTURE OF PKB-BETA (AKT2) COMPLEXED WITH ISOQUINOLINE-5-SULFONIC ACID (2-(2-(4-CHLOROBENZYLOXY) ETHYLAMINO)ETHYL)AMIDE | | Descriptor: | 1,2-ETHANEDIOL, GLYCOGEN SYNTHASE KINASE-3 BETA, ISOQUINOLINE-5-SULFONIC ACID (2-(2-(4-CHLOROBENZYLOXY)ETHYLAMINO)ETHYL)AMIDE, ... | | Authors: | Davies, T.G, Verdonk, M.L, Graham, B, Saalau-Bethell, S, Hamlett, C.C.F, Mchardy, T, Collins, I, Garrett, M.D, Workman, P, Woodhead, S.J, Jhoti, H, Barford, D. | | Deposit date: | 2007-01-11 | | Release date: | 2007-02-13 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | A Structural Comparison of Inhibitor Binding to Pkb, Pka and Pka-Pkb Chimera
J.Mol.Biol., 367, 2007
|
|
5FNU
 
 | | Structure of the Keap1 Kelch domain in complex with a small molecule inhibitor. | | Descriptor: | (3S)-3-(7-methoxy-1-methyl-1H-benzo[d][1,2,3]triazol-5-yl)-3-(4-methyl-3-(((R)-4-methyl-1,1-dioxido-3,4-dihydro-2H-benzo[b][1,4,5]oxathiazepin-2-yl)methyl)phenyl)propanoic acid, CHLORIDE ION, KELCH-LIKE ECH-ASSOCIATED PROTEIN 1 | | Authors: | Davies, T.G, Wixted, W.E, Coyle, J.E, Griffiths-Jones, C, Hearn, K, McMenamin, R, Norton, D, Rich, S.J, Richardson, C, Saxty, G, Willems, H.M.G, Woolford, A.J, Cottom, J.E, Kou, J, Yonchuk, J.G, Feldser, H.G, Sanchez, Y, Foley, J.P, Bolognese, B.J, Logan, G, Podolin, P.L, Yan, H, Callahan, J.F, Heightman, T.D, Kerns, J.K. | | Deposit date: | 2015-11-16 | | Release date: | 2016-04-13 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Mono-Acidic Inhibitors of the Kelch-Like Ech-Associated Protein 1 : Nuclear Factor Erythroid 2-Related Factor 2 (Keap1:Nrf2) Protein-Protein Interaction with High Cell Potency Identified by Fragment-Based Discovery.
J.Med.Chem., 59, 2016
|
|
2JDV
 
 | | Structure of PKA-PKB chimera complexed with A-443654 | | Descriptor: | (2S)-1-(1H-INDOL-3-YL)-3-{[5-(3-METHYL-1H-INDAZOL-5-YL)PYRIDIN-3-YL]OXY}PROPAN-2-AMINE, CAMP-DEPENDENT PROTEIN KINASE, CAMP-DEPENDENT PROTEIN KINASE INHIBITOR ALPHA | | Authors: | Davies, T.G, Verdonk, M.L, Graham, B, Saalau-Bethell, S, Hamlett, C.C.F, Mchardy, T, Collins, I, Garrett, M.D, Workman, P, Woodhead, S.J, Jhoti, H, Barford, D. | | Deposit date: | 2007-01-12 | | Release date: | 2007-02-13 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | A Structural Comparison of Inhibitor Binding to Pkb, Pka and Pka-Pkb Chimera
J.Mol.Biol., 367, 2007
|
|
5FNR
 
 | | Structure of the Keap1 Kelch domain in complex with a small molecule inhibitor. | | Descriptor: | (3S)-3-(4-chlorophenyl)-3-(1-methylbenzotriazol-5-yl)propanoic acid, KELCH-LIKE ECH-ASSOCIATED PROTEIN 1 | | Authors: | Davies, T.G, Wixted, W.E, Coyle, J.E, Griffiths-Jones, C, Hearn, K, McMenamin, R, Norton, D, Rich, S.J, Richardson, C, Saxty, G, Willems, H.M.G, Woolford, A.J, Cottom, J.E, Kou, J, Yonchuk, J.G, Feldser, H.G, Sanchez, Y, Foley, J.P, Bolognese, B.J, Logan, G, Podolin, P.L, Yan, H, Callahan, J.F, Heightman, T.D, Kerns, J.K. | | Deposit date: | 2015-11-16 | | Release date: | 2016-04-13 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Mono-Acidic Inhibitors of the Kelch-Like Ech-Associated Protein 1 : Nuclear Factor Erythroid 2-Related Factor 2 (Keap1:Nrf2) Protein-Protein Interaction with High Cell Potency Identified by Fragment-Based Discovery.
J.Med.Chem., 59, 2016
|
|
2JDR
 
 | | STRUCTURE OF PKB-BETA (AKT2) COMPLEXED WITH THE INHIBITOR A-443654 | | Descriptor: | (2S)-1-(1H-INDOL-3-YL)-3-{[5-(3-METHYL-1H-INDAZOL-5-YL)PYRIDIN-3-YL]OXY}PROPAN-2-AMINE, GLYCOGEN SYNTHASE KINASE-3 BETA, RAC-BETA SERINE/THREONINE-PROTEIN KINASE | | Authors: | Davies, T.G, Verdonk, M.L, Graham, B, Saalau-Bethell, S, Hamlett, C.C.F, McHardy, T, Collins, I, Garrett, M.D, Workman, P, Woodhead, S.J, Jhoti, H, Barford, D. | | Deposit date: | 2007-01-12 | | Release date: | 2007-02-13 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | A Structural Comparison of Inhibitor Binding to Pkb, Pka and Pka-Pkb Chimera
J.Mol.Biol., 367, 2007
|
|
5FNT
 
 | | Structure of the Keap1 Kelch domain in complex with a small molecule inhibitor. | | Descriptor: | (3S)-3-{4-Chloro-3-[(N-methylbenzenesulfonamido) methyl]phenyl}-3-(1-methyl-1H-1,2,3-benzotriazol-5-yl)propanoic acid, CHLORIDE ION, KELCH-LIKE ECH-ASSOCIATED PROTEIN 1 | | Authors: | Davies, T.G, Wixted, W.E, Coyle, J.E, Griffiths-Jones, C, Hearn, K, McMenamin, R, Norton, D, Rich, S.J, Richardson, C, Saxty, G, Willems, H.M.G, Woolford, A.J, Cottom, J.E, Kou, J, Yonchuk, J.G, Feldser, H.G, Sanchez, Y, Foley, J.P, Bolognese, B.J, Logan, G, Podolin, P.L, Yan, H, Callahan, J.F, Heightman, T.D, Kerns, J.K. | | Deposit date: | 2015-11-16 | | Release date: | 2016-04-13 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Mono-Acidic Inhibitors of the Kelch-Like Ech-Associated Protein 1 : Nuclear Factor Erythroid 2-Related Factor 2 (Keap1:Nrf2) Protein-Protein Interaction with High Cell Potency Identified by Fragment-Based Discovery.
J.Med.Chem., 59, 2016
|
|
5FZJ
 
 | | Structure of the Keap1 Kelch domain in complex with a small molecule inhibitor. | | Descriptor: | 2,6-DIMETHYL-4H-PYRANO[3,4-D][1,3]OXAZOL-4-ONE, KELCH-LIKE ECH-ASSOCIATED PROTEIN 1 | | Authors: | Davies, T.G, Wixted, W.E, Coyle, J.E, Griffiths-Jones, C, Hearn, K, McMenamin, R, Norton, D, Rich, S.J, Richardson, C, Saxty, G, Willems, H.M.G, Woolford, A.J, Cottom, J.E, Kou, J, Yonchuk, J.G, Feldser, H.G, Sanchez, Y, Foley, J.P, Bolognese, B.J, Logan, G, Podolin, P.L, Yan, H, Callahan, J.F, Heightman, T.D, Kerns, J.K. | | Deposit date: | 2016-03-14 | | Release date: | 2016-04-13 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Mono-Acidic Inhibitors of the Kelch-Like Ech-Associated Protein 1 : Nuclear Factor Erythroid 2-Related Factor 2 (Keap1:Nrf2) Protein-Protein Interaction with High Cell Potency Identified by Fragment-Based Discovery.
J.Med.Chem., 59, 2016
|
|
5FNS
 
 | | Structure of the Keap1 Kelch domain in complex with a small molecule inhibitor. | | Descriptor: | (3s)-{4-Chloro-3-[(N-methylmethanesulfonamido) methyl]phenyl}-3-(1-methyl-1H-1,2,3-benzotriazol-5-yl) propanoic acid, CHLORIDE ION, KELCH-LIKE ECH-ASSOCIATED PROTEIN 1 | | Authors: | Davies, T.G, Wixted, W.E, Coyle, J.E, Griffiths-Jones, C, Hearn, K, McMenamin, R, Norton, D, Rich, S.J, Richardson, C, Saxty, G, Willems, H.M.G, Woolford, A.J, Cottom, J.E, Kou, J, Yonchuk, J.G, Feldser, H.G, Sanchez, Y, Foley, J.P, Bolognese, B.J, Logan, G, Podolin, P.L, Yan, H, Callahan, J.F, Heightman, T.D, Kerns, J.K. | | Deposit date: | 2015-11-16 | | Release date: | 2016-04-13 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Mono-Acidic Inhibitors of the Kelch-Like Ech-Associated Protein 1 : Nuclear Factor Erythroid 2-Related Factor 2 (Keap1:Nrf2) Protein-Protein Interaction with High Cell Potency Identified by Fragment-Based Discovery.
J.Med.Chem., 59, 2016
|
|
1B2H
 
 | |
1B1H
 
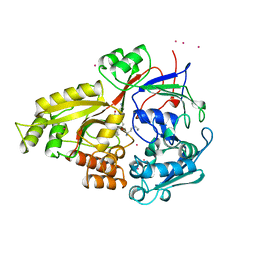 | | OLIGO-PEPTIDE BINDING PROTEIN/TRIPEPTIDE (LYS HPE LYS) COMPLEX | | Descriptor: | PROTEIN (LYS HPE LYS), PROTEIN (OLIGO-PEPTIDE BINDING PROTEIN), URANYL (VI) ION | | Authors: | Davies, T.G, Tame, J.R.H. | | Deposit date: | 1998-11-10 | | Release date: | 1998-11-18 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Relating structure to thermodynamics: the crystal structures and binding affinity of eight OppA-peptide complexes.
Protein Sci., 8, 1999
|
|
1B3H
 
 | |
1B5H
 
 | |
1B4H
 
 | |
1B0H
 
 | |
1B7H
 
 | |
1B6H
 
 | |
1E9H
 
 | | Thr 160 phosphorylated CDK2 - Human cyclin A3 complex with the inhibitor indirubin-5-sulphonate bound | | Descriptor: | 2',3-DIOXO-1,1',2',3-TETRAHYDRO-2,3'-BIINDOLE-5'-SULFONIC ACID, CELL DIVISION PROTEIN KINASE 2, CYCLIN A3 | | Authors: | Davies, T.G, Tunnah, P, Noble, M.E.M, Endicott, J.A. | | Deposit date: | 2000-10-16 | | Release date: | 2001-10-11 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Inhibitor Binding to Active and Inactive Cdk2: The Crystal Structure of Cdk2-Cyclin A/Indirubin-5-Sulphonate
Structure, 9, 2001
|
|
