6TCI
 
 | |
1NK1
 
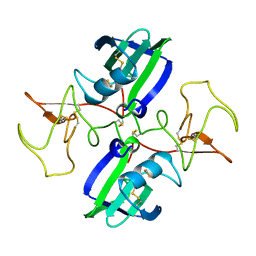 | | NK1 FRAGMENT OF HUMAN HEPATOCYTE GROWTH FACTOR/SCATTER FACTOR (HGF/SF) AT 2.5 ANGSTROM RESOLUTION | | Descriptor: | PROTEIN (HEPATOCYTE GROWTH FACTOR PRECURSOR) | | Authors: | Chirgadze, D.Y, Hepple, J.P, Zhou, H, Byrd, R.A, Blundell, T.L, Gherardi, E. | | Deposit date: | 1998-08-20 | | Release date: | 1999-01-13 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of the NK1 fragment of HGF/SF suggests a novel mode for growth factor dimerization and receptor binding.
Nat.Struct.Biol., 6, 1999
|
|
1U41
 
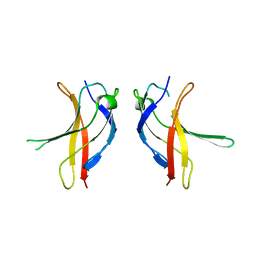 | | Crystal structure of YLGV mutant of dimerisation domain of NF-kB p50 transcription factor | | Descriptor: | Nuclear factor NF-kappa-B p105 subunit | | Authors: | Chirgadze, D.Y, Demydchuk, M, Becker, M, Moran, S, Paoli, M. | | Deposit date: | 2004-07-23 | | Release date: | 2004-08-17 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.202 Å) | | Cite: | Snapshot of Protein Structure Evolution Reveals Conservation of Functional Dimerization through Intertwined Folding
Structure, 12, 2004
|
|
1U3Y
 
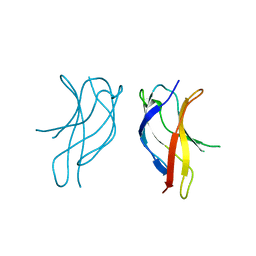 | | Crystal structure of ILAC mutant of dimerisation domain of NF-kB p50 transcription factor | | Descriptor: | Nuclear factor NF-kappa-B p105 subunit | | Authors: | Chirgadze, D.Y, Demydchuk, M, Becker, M, Moran, S, Paoli, M. | | Deposit date: | 2004-07-22 | | Release date: | 2004-08-17 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.901 Å) | | Cite: | Snapshot of Protein Structure Evolution Reveals Conservation of Functional Dimerization through Intertwined Folding
Structure, 12, 2004
|
|
1U42
 
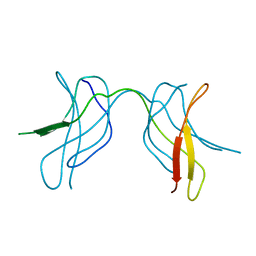 | | Crystal structure of MLAM mutant of dimerisation domain of NF-kB p50 transcription factor | | Descriptor: | Nuclear factor NF-kappa-B p105 subunit | | Authors: | Chirgadze, D.Y, Demydchuk, M, Becker, M, Moran, S, Paoli, M. | | Deposit date: | 2004-07-23 | | Release date: | 2004-08-17 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.699 Å) | | Cite: | Snapshot of Protein Structure Evolution Reveals Conservation of Functional Dimerization through Intertwined Folding
Structure, 12, 2004
|
|
1U3Z
 
 | | Crystal structure of MLAC mutant of dimerisation domain of NF-kB p50 transcription factor | | Descriptor: | Nuclear factor NF-kappa-B p105 subunit | | Authors: | Chirgadze, D.Y, Demydchuk, M, Becker, M, Moran, S, Paoli, M. | | Deposit date: | 2004-07-23 | | Release date: | 2004-08-17 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Snapshot of Protein Structure Evolution Reveals Conservation of Functional Dimerization through Intertwined Folding
Structure, 12, 2004
|
|
1U3J
 
 | | Crystal structure of MLAV mutant of dimerisation domain of NF-kB p50 transcription factor | | Descriptor: | Nuclear factor NF-kappa-B p105 subunit | | Authors: | Chirgadze, D.Y, Demydchuk, M, Becker, M, Moran, S, Paoli, M. | | Deposit date: | 2004-07-22 | | Release date: | 2004-08-17 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.901 Å) | | Cite: | Snapshot of Protein Structure Evolution Reveals Conservation of Functional Dimerization through Intertwined Folding
Structure, 12, 2004
|
|
1U36
 
 | | Crystal structure of WLAC mutant of dimerisation domain of NF-kB p50 transcription factor | | Descriptor: | Nuclear factor NF-kappa-B p105 subunit | | Authors: | Chirgadze, D.Y, Demydchuk, M, Becker, M, Moran, S, Paoli, M. | | Deposit date: | 2004-07-21 | | Release date: | 2004-08-17 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Snapshot of Protein Structure Evolution Reveals Conservation of Functional Dimerization through Intertwined Folding
Structure, 12, 2004
|
|
5JIP
 
 | | Crystal structure of the Clostridium perfringens spore cortex lytic enzyme SleM | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Cortical-lytic enzyme, MAGNESIUM ION | | Authors: | Chirgadze, D.Y, Christie, G, Ustok, F.I, Al-Riyami, B, Stott, K. | | Deposit date: | 2016-04-22 | | Release date: | 2016-08-10 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The crystal structure of Clostridium perfringens SleM, a muramidase involved in cortical hydrolysis during spore germination.
Proteins, 84, 2016
|
|
3KX8
 
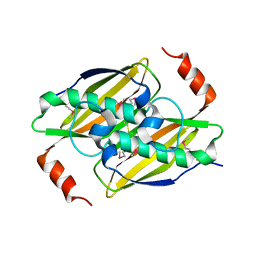 | | Structural basis of the activity and substrate specificity of the fluoroacetyl-CoA thioesterase FlK | | Descriptor: | fluoroacetyl-CoA thioesterase | | Authors: | Chirgadze, D.Y, Dias, M.V.B, Huang, F, Tosin, M, Spiteller, D, Valentine, E.F, Leadlay, P.F, Spencer, J.B, Blundell, T.L. | | Deposit date: | 2009-12-02 | | Release date: | 2010-04-28 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structural basis for the activity and substrate specificity of fluoroacetyl-CoA thioesterase FlK.
J.Biol.Chem., 285, 2010
|
|
3G04
 
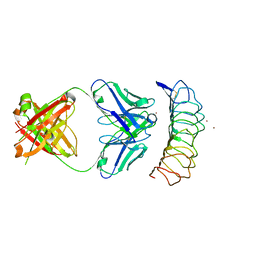 | | Crystal structure of the TSH receptor in complex with a thyroid-stimulating autoantibody | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, HUMAN THYROID STIMULATING AUTOANTIBODY M22 HEAVY CHAIN, HUMAN THYROID STIMULATING AUTOANTIBODY M22 LIGHT CHAIN, ... | | Authors: | Sanders, J, Chirgadze, D.Y, Sanders, P, Baker, S, Sullivan, A, Bhardwaja, A, Bolton, J, Reeve, M, Nakatake, N, Evans, M, Richards, T, Powell, M, Miguel, R.N, Blundell, T.L, Furmaniak, J, Smith, B.R. | | Deposit date: | 2009-01-27 | | Release date: | 2009-08-04 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Crystal structure of the TSH receptor in complex with a thyroid-stimulating autoantibody
Thyroid, 17, 2007
|
|
8B0J
 
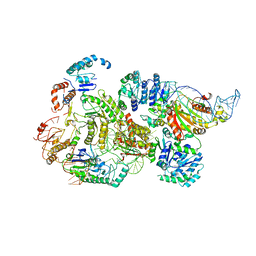 | | CryoEM structure of bacterial RNaseE.RapZ.GlmZ complex central to the control of cell envelope biogenesis | | Descriptor: | GlmZ small RNA, RNase adapter protein RapZ, Ribonuclease E | | Authors: | Islam, M.S, Hardwick, H.W, Chirgadze, D.Y, Luisi, B.F. | | Deposit date: | 2022-09-07 | | Release date: | 2022-10-05 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.99 Å) | | Cite: | Structure of a bacterial ribonucleoprotein complex central to the control of cell envelope biogenesis.
Embo J., 42, 2023
|
|
8B0I
 
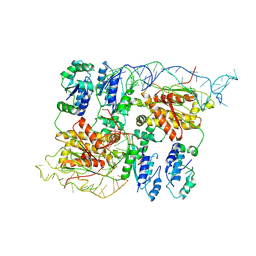 | | CryoEM structure of bacterial RapZ.GlmZ complex central to the control of cell envelope biogenesis | | Descriptor: | GlmZ small regulatory RNA, RNase adapter protein RapZ | | Authors: | Islam, M.S, Hardwick, H.W, Chirgadze, D.Y, Luisi, B.F. | | Deposit date: | 2022-09-07 | | Release date: | 2022-10-05 | | Last modified: | 2025-07-09 | | Method: | ELECTRON MICROSCOPY (4.28 Å) | | Cite: | Structure of a bacterial ribonucleoprotein complex central to the control of cell envelope biogenesis.
Embo J., 42, 2023
|
|
8PET
 
 | | Cryo-EM structure of the full-length human alpha1beta3 GABA(A) receptor (babba arrangement) in complex with nanobody Nb25, expressed with a 5-fold excess of beta3 over alpha1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, DECANE, Gamma-aminobutyric acid receptor subunit alpha-1, ... | | Authors: | Sente, A, Malinauskas, T, Naydenova, K, Hardwick, S.W, Chirgadze, D.Y, Aricescu, A.R. | | Deposit date: | 2023-06-14 | | Release date: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Subunit abundance shapes GABAA receptor stoichiometry
To Be Published
|
|
6ZFP
 
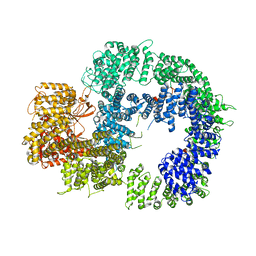 | | Cryo-EM structure of DNA-PKcs (State 2) | | Descriptor: | DNA-dependent protein kinase catalytic subunit,DNA-PKcs,DNA-PKcs | | Authors: | Chaplin, A.K, Hardwick, S.W, Chirgadze, D.Y, Blundell, T.L. | | Deposit date: | 2020-06-17 | | Release date: | 2020-10-21 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (3.24 Å) | | Cite: | Dimers of DNA-PK create a stage for DNA double-strand break repair.
Nat.Struct.Mol.Biol., 28, 2021
|
|
6ZHE
 
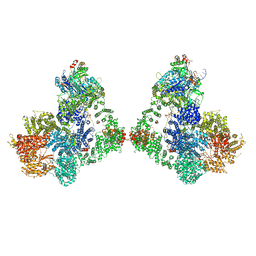 | | Cryo-EM structure of DNA-PK dimer | | Descriptor: | DNA (25-MER), DNA (26-MER), DNA (27-MER), ... | | Authors: | Chaplin, A.K, Hardwick, S.W, Chirgadze, D.Y, Blundell, T.L. | | Deposit date: | 2020-06-23 | | Release date: | 2020-10-21 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (7.24 Å) | | Cite: | Dimers of DNA-PK create a stage for DNA double-strand break repair.
Nat.Struct.Mol.Biol., 28, 2021
|
|
6ZH2
 
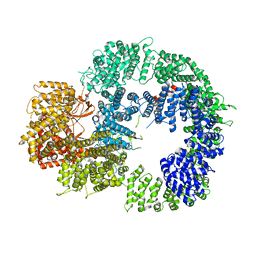 | | Cryo-EM structure of DNA-PKcs (State 1) | | Descriptor: | DNA-dependent protein kinase catalytic subunit,DNA-dependent protein kinase catalytic subunit,DNA-dependent protein kinase catalytic subunit,DNA-dependent protein kinase catalytic subunit,DNA-dependent protein kinase catalytic subunit,DNA-dependent protein kinase catalytic subunit,DNA-dependent protein kinase catalytic subunit,DNA-dependent protein kinase catalytic subunit,DNA-PKcs | | Authors: | Chaplin, A.K, Hardwick, S.W, Chirgadze, D.Y, Blundell, T.L. | | Deposit date: | 2020-06-20 | | Release date: | 2020-10-21 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (3.92 Å) | | Cite: | Dimers of DNA-PK create a stage for DNA double-strand break repair.
Nat.Struct.Mol.Biol., 28, 2021
|
|
6ZH4
 
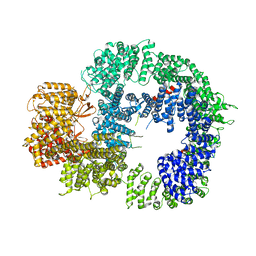 | | Cryo-EM structure of DNA-PKcs (State 3) | | Descriptor: | DNA-dependent protein kinase catalytic subunit,DNA-PKcs | | Authors: | Chaplin, A.K, Hardwick, S.W, Chirgadze, D.Y, Blundell, T.L. | | Deposit date: | 2020-06-20 | | Release date: | 2020-10-21 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (3.62 Å) | | Cite: | Dimers of DNA-PK create a stage for DNA double-strand break repair.
Nat.Struct.Mol.Biol., 28, 2021
|
|
6ZH6
 
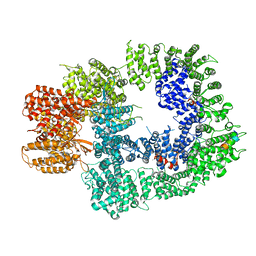 | | Cryo-EM structure of DNA-PKcs:Ku80ct194 | | Descriptor: | DNA-dependent protein kinase catalytic subunit,DNA-PKcs, X-ray repair cross-complementing protein 5 | | Authors: | Chaplin, A.K, Hardwick, S.W, Chirgadze, D.Y, Blundell, T.L. | | Deposit date: | 2020-06-21 | | Release date: | 2020-10-21 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (3.93 Å) | | Cite: | Dimers of DNA-PK create a stage for DNA double-strand break repair.
Nat.Struct.Mol.Biol., 28, 2021
|
|
3COW
 
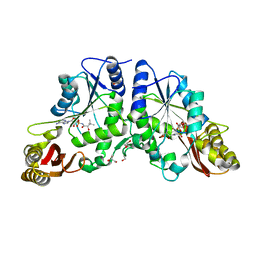 | | Crystal Structure of Mycobacterium Tuberculosis Pantothenate Synthetase at 1.8 Ang resolution- in complex with sulphonamide inhibitor 2 | | Descriptor: | 5'-O-{[(2R)-2-hydroxy-3,3-dimethylbutanoyl]sulfamoyl}adenosine, ETHANOL, GLYCEROL, ... | | Authors: | Ciulli, A, Chirgadze, D.Y, Blundell, T.L, Abell, C. | | Deposit date: | 2008-03-29 | | Release date: | 2008-10-07 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Inhibition of Mycobacterium tuberculosis pantothenate synthetase by analogues of the reaction intermediate.
Chembiochem, 9, 2008
|
|
6XTY
 
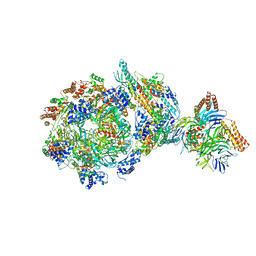 | | CryoEM structure of human CMG bound to AND-1 (CMGA) | | Descriptor: | Cell division control protein 45 homolog, DNA replication complex GINS protein PSF1, DNA replication complex GINS protein PSF2, ... | | Authors: | Rzechorzek, N.J, Pellegrini, L, Chirgadze, D.Y, Hardwick, S.W. | | Deposit date: | 2020-01-16 | | Release date: | 2020-05-27 | | Last modified: | 2025-07-09 | | Method: | ELECTRON MICROSCOPY (6.77 Å) | | Cite: | CryoEM structures of human CMG-ATP gamma S-DNA and CMG-AND-1 complexes.
Nucleic Acids Res., 48, 2020
|
|
2J66
 
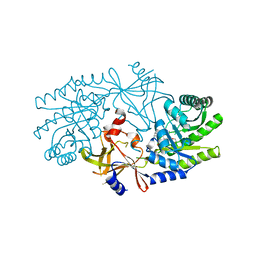 | | Structural characterisation of BtrK decarboxylase from butirosin biosynthesis | | Descriptor: | 1,2-ETHANEDIOL, BTRK, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Popovic, B, Li, Y, Chirgadze, D.Y, Blundell, T.L, Spencer, J.B. | | Deposit date: | 2006-09-26 | | Release date: | 2006-09-28 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structural Characterisation of Btrk Decarboxylase from Bacillus Circulans Butirosin Biosynthesis
To be Published
|
|
7ZYG
 
 | | CryoEM structure of Ku heterodimer bound to DNA, PAXX and XLF | | Descriptor: | DNA, Non-homologous end-joining factor 1, PHOSPHATE ION, ... | | Authors: | Hardwick, S.W, Kefala-Stavridi, A, Chirgadze, D.Y, Blundell, T.L, Chaplin, A.K. | | Deposit date: | 2022-05-24 | | Release date: | 2023-06-07 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (2.68 Å) | | Cite: | PAXX binding to the NHEJ machinery explains functional redundancy with XLF.
Sci Adv, 9, 2023
|
|
1URF
 
 | | HR1b domain from PRK1 | | Descriptor: | PROTEIN KINASE C-LIKE 1 | | Authors: | Owen, D, Lowe, P.N, Nietlispach, D, Brosnan, C.E, Chirgadze, D.Y, Parker, P.J, Blundell, T.L, Mott, H.R. | | Deposit date: | 2003-10-29 | | Release date: | 2003-11-06 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Molecular Dissection of the Interaction between the Small G Proteins Rac1 and Rhoa and Protein Kinase C-Related Kinase 1 (Prk1)
J.Biol.Chem., 278, 2003
|
|
3E4R
 
 | | Crystal structure of the alkanesulfonate binding protein (SsuA) from the phytopathogenic bacteria Xanthomonas axonopodis pv. citri bound to HEPES | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Nitrate transport protein | | Authors: | Balan, A, Araujo, F.T, Sanches, M, Chirgadze, D.Y, Blundell, T.B, Barbosa, J.A.R.G. | | Deposit date: | 2008-08-12 | | Release date: | 2008-09-23 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Crystal structure of the alkanesulfonate binding protein (SsuA) from the phytopathogenic bacteria Xanthomonas axonopodis pv. citri bound to HEPES
To be Published
|
|
