1Z9E
 
 | | Solution structure of the HIV-1 integrase-binding domain in LEDGF/p75 | | Descriptor: | PC4 and SFRS1 interacting protein 2 | | Authors: | Cherepanov, P, Sun, Z.-Y.J, Rahman, S, Maertens, G, Wagner, G, Engelman, A. | | Deposit date: | 2005-04-01 | | Release date: | 2005-05-17 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the HIV-1 integrase-binding domain in LEDGF/p75
Nat.Struct.Mol.Biol., 12, 2005
|
|
2B4J
 
 | | Structural basis for the recognition between HIV-1 integrase and LEDGF/p75 | | Descriptor: | GLYCEROL, Integrase (IN), PC4 and SFRS1 interacting protein, ... | | Authors: | Cherepanov, P, Ambrosio, A.L, Rahman, S, Ellenberger, T, Engelman, A. | | Deposit date: | 2005-09-24 | | Release date: | 2005-10-25 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Structural basis for the recognition between HIV-1 integrase and transcriptional coactivator p75
Proc.Natl.Acad.Sci.Usa, 102, 2005
|
|
6GX9
 
 | | Crystal structure of the TNPO3 - CPSF6 RSLD complex | | Descriptor: | BENZAMIDINE, BICINE, Cleavage and polyadenylation specificity factor subunit 6, ... | | Authors: | Cherepanov, P, Cook, N. | | Deposit date: | 2018-06-26 | | Release date: | 2019-03-13 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Differential role for phosphorylation in alternative polyadenylation function versus nuclear import of SR-like protein CPSF6.
Nucleic Acids Res., 47, 2019
|
|
6RWO
 
 | | SIVrcm intasome (Q148H/G140S) in complex with bictegravir | | Descriptor: | Bictegravir, CHLORIDE ION, DNA (5'-D(*AP*AP*CP*TP*GP*GP*TP*AP*GP*AP*GP*AP*TP*TP*TP*TP*TP*CP*TP*TP*AP*GP*C)-3'), ... | | Authors: | Cherepanov, P, Nans, A, Cook, N. | | Deposit date: | 2019-06-05 | | Release date: | 2020-02-05 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (3.05 Å) | | Cite: | Structural basis of second-generation HIV integrase inhibitor action and viral resistance.
Science, 367, 2020
|
|
6RWL
 
 | | SIVrcm intasome | | Descriptor: | DNA (5'-D(*AP*AP*CP*TP*GP*GP*TP*AP*GP*AP*GP*AP*TP*TP*TP*TP*TP*CP*TP*TP*AP*GP*C)-3'), DNA (5'-D(P*GP*CP*TP*AP*AP*GP*AP*AP*AP*AP*AP*TP*CP*TP*CP*TP*AP*CP*CP*A)-3'), Pol protein, ... | | Authors: | Cherepanov, P, Nans, A, Cook, N. | | Deposit date: | 2019-06-05 | | Release date: | 2020-02-05 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (3.36 Å) | | Cite: | Structural basis of second-generation HIV integrase inhibitor action and viral resistance.
Science, 367, 2020
|
|
6RWM
 
 | | SIVrcm intasome in complex with bictegravir | | Descriptor: | Bictegravir, CHLORIDE ION, DNA (5'-D(*AP*AP*CP*TP*GP*GP*TP*AP*GP*AP*GP*AP*TP*TP*TP*TP*TP*CP*TP*TP*AP*GP*C)-3'), ... | | Authors: | Cherepanov, P, Nans, A, Cook, N. | | Deposit date: | 2019-06-05 | | Release date: | 2020-02-05 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (2.81 Å) | | Cite: | Structural basis of second-generation HIV integrase inhibitor action and viral resistance.
Science, 367, 2020
|
|
6RWN
 
 | | SIVrcm intasome in complex with dolutegravir | | Descriptor: | (4R,12aS)-N-(2,4-difluorobenzyl)-7-hydroxy-4-methyl-6,8-dioxo-3,4,6,8,12,12a-hexahydro-2H-pyrido[1',2':4,5]pyrazino[2,1-b][1,3]oxazine-9-carboxamide, CHLORIDE ION, DNA (5'-D(*AP*AP*CP*TP*GP*GP*TP*AP*GP*AP*GP*AP*TP*TP*TP*TP*TP*CP*TP*TP*AP*GP*C)-3'), ... | | Authors: | Cherepanov, P, Nans, A, Cook, N. | | Deposit date: | 2019-06-05 | | Release date: | 2020-02-05 | | Last modified: | 2024-07-10 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural basis of second-generation HIV integrase inhibitor action and viral resistance.
Science, 367, 2020
|
|
5FRO
 
 | | Crystal structure of the Prototype Foamy Virus (PFV) intasome in complex with magnesium and the INSTI XZ446 (compound 4f) | | Descriptor: | 4-azanyl-N-[[2,4-bis(fluoranyl)phenyl]methyl]-1-oxidanyl-2-oxidanylidene-6-[2-(phenylsulfonyl)ethyl]-1,8-naphthyridine-3-carboxamide, 5'-D(*AP*TP*TP*GP*TP*CP*AP*TP*GP*GP*AP*AP*TP*TP *TP*CP*GP*CP*A)-3', 5'-D(*TP*GP*CP*GP*AP*AP*AP*TP*TP*CP*CP*AP*TP*GP *AP*CP*A)-3', ... | | Authors: | Maskell, D.P, Pye, V.E, Cherepanov, P. | | Deposit date: | 2015-12-18 | | Release date: | 2016-02-17 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.67 Å) | | Cite: | HIV-1 Integrase Strand Transfer Inhibitors with Reduced Susceptibility to Drug Resistant Mutant Integrases.
Acs Chem.Biol., 11, 2016
|
|
5M0R
 
 | | Cryo-EM reconstruction of the maedi-visna virus (MVV) strand transfer complex | | Descriptor: | integrase, tDNA, vDNA, ... | | Authors: | Pye, V.E, Ballandras-Colas, A, Maskell, D, Locke, J, Kotecha, A, Costa, A, Cherepanov, P. | | Deposit date: | 2016-10-05 | | Release date: | 2017-01-18 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (8.2 Å) | | Cite: | A supramolecular assembly mediates lentiviral DNA integration.
Science, 355, 2017
|
|
7B62
 
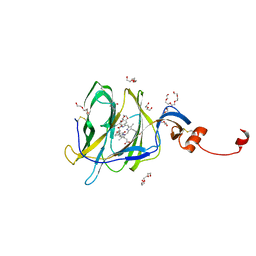 | | Crystal structure of SARS-CoV-2 spike protein N-terminal domain in complex with biliverdin | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, BILIVERDINE IX ALPHA, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Pye, V.E, Rosa, A, Roustan, C, Cherepanov, P. | | Deposit date: | 2020-12-07 | | Release date: | 2021-04-28 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | SARS-CoV-2 can recruit a heme metabolite to evade antibody immunity.
Sci Adv, 7, 2021
|
|
8POE
 
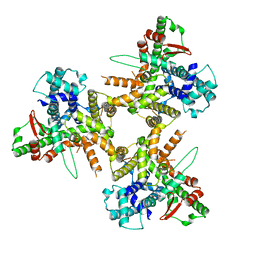 | | Structure of tissue-specific lipid scramblase ATG9B homotrimer, refined with C3 symmetry applied | | Descriptor: | Autophagy-related protein 9B | | Authors: | Chiduza, G.N, Pye, V.E, Tooze, S.A, Cherepanov, P. | | Deposit date: | 2023-07-04 | | Release date: | 2023-11-15 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | ATG9B is a tissue-specific homotrimeric lipid scramblase that can compensate for ATG9A.
Autophagy, 20, 2024
|
|
5MEC
 
 | |
5LLJ
 
 | |
6YA6
 
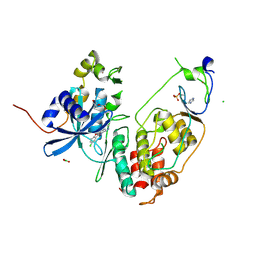 | | Minimal construct of Cdc7-Dbf4 bound to XL413 | | Descriptor: | 8-chloro-2-[(2S)-pyrrolidin-2-yl][1]benzofuro[3,2-d]pyrimidin-4(3H)-one, BORIC ACID, CHLORIDE ION, ... | | Authors: | Dick, S.D, Cherepanov, P. | | Deposit date: | 2020-03-11 | | Release date: | 2020-05-27 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.44 Å) | | Cite: | Structural Basis for the Activation and Target Site Specificity of CDC7 Kinase.
Structure, 28, 2020
|
|
8CBR
 
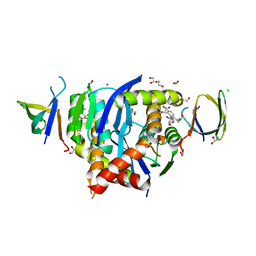 | | HIV-1 Integrase Catalytic Core Domain and C-Terminal Domain in Complex with Allosteric Integrase Inhibitor BDM-2 | | Descriptor: | (2S)-2-[3-cyclopropyl-2-(3,4-dihydro-2H-chromen-6-yl)-6-methyl-phenyl]-2-[(2-methylpropan-2-yl)oxy]ethanoic acid, 1,2-ETHANEDIOL, CHLORIDE ION, ... | | Authors: | Singer, M.R, Pye, V.E, Yu, Z, Cherepanov, P. | | Deposit date: | 2023-01-25 | | Release date: | 2023-06-07 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Biological and Structural Analyses of New Potent Allosteric Inhibitors of HIV-1 Integrase.
Antimicrob.Agents Chemother., 67, 2023
|
|
8CBU
 
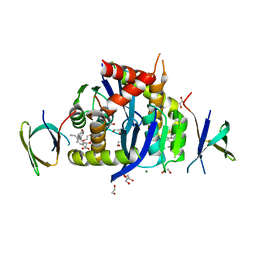 | | HIV-1 Integrase Catalytic Core Domain and C-Terminal Domain in Complex with Allosteric Integrase Inhibitor MUT884 | | Descriptor: | (2S)-2-[3-cyclopropyl-6-methyl-2-(5-methyl-3,4-dihydro-2H-chromen-6-yl)phenyl]-2-cyclopropyloxy-ethanoic acid, 1,2-ETHANEDIOL, CHLORIDE ION, ... | | Authors: | Singer, M.R, Pye, V.E, Yu, Z, Cherepanov, P. | | Deposit date: | 2023-01-25 | | Release date: | 2023-06-07 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.44 Å) | | Cite: | Biological and Structural Analyses of New Potent Allosteric Inhibitors of HIV-1 Integrase.
Antimicrob.Agents Chemother., 67, 2023
|
|
8CBV
 
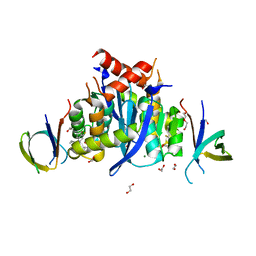 | | HIV-1 Integrase Catalytic Core Domain and C-Terminal Domain in Complex with Allosteric Integrase Inhibitor MUT916 | | Descriptor: | (2~{S})-2-[3-cyclopropyl-2-(8-fluoranyl-5-methyl-3,4-dihydro-2~{H}-chromen-6-yl)-6-methyl-phenyl]-2-cyclopropyloxy-ethanoic acid, 1,2-ETHANEDIOL, CHLORIDE ION, ... | | Authors: | Singer, M.R, Pye, V.E, Yu, Z, Cherepanov, P. | | Deposit date: | 2023-01-25 | | Release date: | 2023-06-07 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Biological and Structural Analyses of New Potent Allosteric Inhibitors of HIV-1 Integrase.
Antimicrob.Agents Chemother., 67, 2023
|
|
8CBT
 
 | | HIV-1 Integrase Catalytic Core Domain and C-Terminal Domain in Complex with Allosteric Integrase Inhibitor MUT872 | | Descriptor: | (2~{S})-2-[3-cyclopropyl-2-(3,4-dihydro-2~{H}-chromen-6-yl)-6-methyl-phenyl]-2-cyclopropyloxy-ethanoic acid, 1,2-ETHANEDIOL, CHLORIDE ION, ... | | Authors: | Singer, M.R, Pye, V.E, Yu, Z, Cherepanov, P. | | Deposit date: | 2023-01-25 | | Release date: | 2023-06-07 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | Biological and Structural Analyses of New Potent Allosteric Inhibitors of HIV-1 Integrase.
Antimicrob.Agents Chemother., 67, 2023
|
|
8CBS
 
 | | HIV-1 Integrase Catalytic Core Domain and C-Terminal Domain in Complex with Allosteric Integrase Inhibitor MUT871 | | Descriptor: | (2~{S})-2-[3-cyclopropyl-6-methyl-2-(5-methyl-3,4-dihydro-2~{H}-chromen-6-yl)phenyl]-2-[(2-methylpropan-2-yl)oxy]ethanoic acid, 1,2-ETHANEDIOL, CHLORIDE ION, ... | | Authors: | Singer, M.R, Pye, V.E, Yu, Z, Cherepanov, P. | | Deposit date: | 2023-01-25 | | Release date: | 2023-06-07 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Biological and Structural Analyses of New Potent Allosteric Inhibitors of HIV-1 Integrase.
Antimicrob.Agents Chemother., 67, 2023
|
|
5FRM
 
 | | Crystal structure of the Prototype Foamy Virus (PFV) intasome in complex with magnesium and the INSTI XZ384 (compound 4a) | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 4-azanylidene-N-[[2,4-bis(fluoranyl)phenyl]methyl]-1-oxidanyl-2-oxidanylidene-1,8-naphthyridine-3-carboxamide, 5'-D(*AP*TP*TP*GP*TP*CP*AP*TP*GP*GP*AP*AP*TP*TP *TP*CP*GP*CP*A)-3', ... | | Authors: | Maskell, D.P, Pye, V.E, Cherepanov, P. | | Deposit date: | 2015-12-18 | | Release date: | 2016-02-17 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.58 Å) | | Cite: | HIV-1 Integrase Strand Transfer Inhibitors with Reduced Susceptibility to Drug Resistant Mutant Integrases.
Acs Chem.Biol., 11, 2016
|
|
3HPH
 
 | | Closed tetramer of Visna virus integrase (residues 1-219) in complex with LEDGF IBD | | Descriptor: | GLYCEROL, Integrase, PC4 and SFRS1-interacting protein, ... | | Authors: | Hare, S, Wang, J, Cherepanov, P. | | Deposit date: | 2009-06-04 | | Release date: | 2009-07-28 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.64 Å) | | Cite: | Structural basis for functional tetramerization of lentiviral integrase
Plos Pathog., 5, 2009
|
|
7Z1Z
 
 | | MVV strand transfer complex (STC) intasome in complex with LEDGF/p75 at 3.5 A resolution | | Descriptor: | DNA (37-MER), DNA (5'-D(*GP*CP*TP*GP*CP*GP*AP*GP*AP*TP*CP*CP*GP*CP*TP*CP*CP*GP*GP*TP*G)-3'), DNA (5'-D(P*TP*TP*GP*AP*TP*TP*AP*GP*GP*GP*TP*G)-3'), ... | | Authors: | Pye, V.E, Ballandras-Colas, A, Cherepanov, P. | | Deposit date: | 2022-02-25 | | Release date: | 2022-05-11 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Multivalent interactions essential for lentiviral integrase function.
Nat Commun, 13, 2022
|
|
7ZBU
 
 | | CryoEM structure of SARS-CoV-2 spike monomer in complex with neutralising antibody P008_60 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 3-[5-[(4-ethyl-3-methyl-5-oxidanylidene-pyrrol-2-yl)methyl]-2-[[5-[(3-ethyl-4-methyl-5-oxidanylidene-pyrrol-2-yl)methyl]-3-(3-hydroxy-3-oxopropyl)-4-methyl-1H-pyrrol-2-yl]methyl]-4-methyl-1H-pyrrol-3-yl]propanoic acid, P008_60 antibody, ... | | Authors: | Rosa, A, Pye, V.E, Cronin, N, Cherepanov, P. | | Deposit date: | 2022-03-24 | | Release date: | 2022-08-17 | | Last modified: | 2022-08-31 | | Method: | ELECTRON MICROSCOPY (4.31 Å) | | Cite: | A neutralizing epitope on the SD1 domain of SARS-CoV-2 spike targeted following infection and vaccination.
Cell Rep, 40, 2022
|
|
5T3A
 
 | |
7U32
 
 | | MVV cleaved synaptic complex (CSC) intasome at 3.4 A resolution | | Descriptor: | CALCIUM ION, DNA EV272, DNA EV273, ... | | Authors: | Shan, Z, Pye, V.E, Cherepanov, P, Lyumkis, D. | | Deposit date: | 2022-02-25 | | Release date: | 2022-05-11 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (3.46 Å) | | Cite: | Multivalent interactions essential for lentiviral integrase function.
Nat Commun, 13, 2022
|
|
