1CEM
 
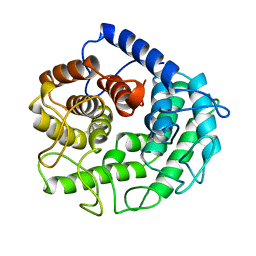 | | ENDOGLUCANASE A (CELA) CATALYTIC CORE, RESIDUES 33-395 | | Descriptor: | CELLULASE CELA (1,4-BETA-D-GLUCAN-GLUCANOHYDROLASE) | | Authors: | Alzari, P.M. | | Deposit date: | 1995-12-04 | | Release date: | 1997-01-11 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | The crystal structure of endoglucanase CelA, a family 8 glycosyl hydrolase from Clostridium thermocellum.
Structure, 4, 1996
|
|
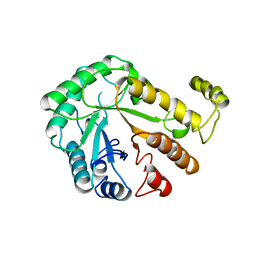
1CEC
 
 | |
6CAC
 
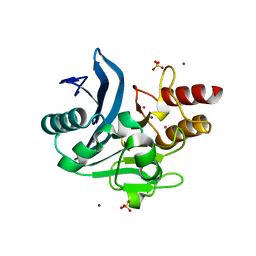 | | Crystal structure of NDM-1 metallo-beta-lactamase harboring an insertion of a Pro residue in L3 loop | | Descriptor: | CADMIUM ION, CALCIUM ION, COBALT (II) ION, ... | | Authors: | Alzari, P.M, Giannini, E, Palacios, A, Mojica, M, Bonomo, R, Llarrull, L, Vila, A. | | Deposit date: | 2018-01-30 | | Release date: | 2018-10-17 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | The Reaction Mechanism of Metallo-beta-Lactamases Is Tuned by the Conformation of an Active-Site Mobile Loop.
Antimicrob. Agents Chemother., 63, 2019
|
|
1CLC
 
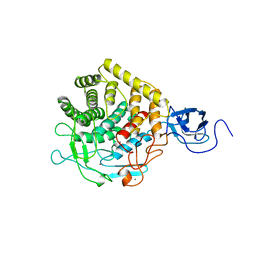 | |
1XYZ
 
 | |
1AOH
 
 | |
1HHL
 
 | |
1GHL
 
 | |
1F4X
 
 | |
1F4W
 
 | |
1F4Y
 
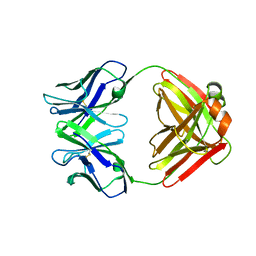 | | CRYSTAL STRUCTURE OF AN ANTI-CARBOHYDRATE ANTIBODY DIRECTED AGAINST VIBRIO CHOLERAE O1 IN COMPLEX WITH ANTIGEN | | Descriptor: | 4,6-dideoxy-4-{[(2R)-2,4-dihydroxybutanoyl]amino}-2-O-methyl-alpha-D-mannopyranose-(1-2)-methyl 4,6-dideoxy-4-{[(2R)-2,4-dihydroxybutanoyl]amino}-alpha-D-mannopyranoside, ANTIBODY S-20-4, FAB FRAGMENT, ... | | Authors: | Alzari, P.M, Souchon, H. | | Deposit date: | 2000-06-10 | | Release date: | 2000-08-02 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of an anti-carbohydrate antibody directed against Vibrio cholerae O1 in complex with antigen: molecular basis for serotype specificity.
Proc.Natl.Acad.Sci.USA, 97, 2000
|
|
2GEK
 
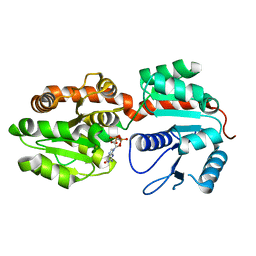 | | Crystal Structure of phosphatidylinositol mannosyltransferase (PimA) from Mycobacterium smegmatis in complex with GDP | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, PHOSPHATIDYLINOSITOL MANNOSYLTRANSFERASE (PimA) | | Authors: | Guerin, M.E, Buschiazzo, A, Kordulakova, J, Jackson, M, Alzari, P.M. | | Deposit date: | 2006-03-20 | | Release date: | 2007-04-03 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Molecular recognition and interfacial catalysis by the essential phosphatidylinositol mannosyltransferase PimA from mycobacteria.
J.Biol.Chem., 282, 2007
|
|
2GEJ
 
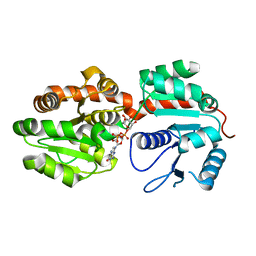 | | Crystal Structure of phosphatidylinositol mannosyltransferase (PimA) from Mycobacterium smegmatis in complex with GDP-Man | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE-ALPHA-D-MANNOSE, PHOSPHATIDYLINOSITOL MANNOSYLTRANSFERASE (PimA) | | Authors: | Guerin, M.E, Buschiazzo, A, Kordulakova, J, Jackson, M, Alzari, P.M. | | Deposit date: | 2006-03-20 | | Release date: | 2007-04-03 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Molecular recognition and interfacial catalysis by the essential phosphatidylinositol mannosyltransferase PimA from mycobacteria.
J.Biol.Chem., 282, 2007
|
|
7A1D
 
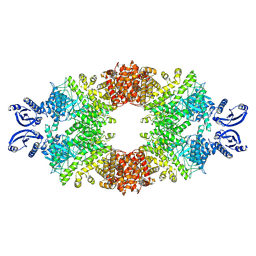 | | Cryo-EM map of the large glutamate dehydrogenase composed of 180 kDa subunits from Mycobacterium smegmatis (open conformation) | | Descriptor: | NAD-specific glutamate dehydrogenase | | Authors: | Lazaro, M, Melero, R, Huet, C, Lopez-Alonso, J.P, Delgado, S, Dodu, A, Bruch, E.M, Abriata, L.A, Alzari, P.M, Valle, M, Lisa, M.N. | | Deposit date: | 2020-08-12 | | Release date: | 2021-06-09 | | Last modified: | 2025-05-14 | | Method: | ELECTRON MICROSCOPY (4.19 Å) | | Cite: | 3D architecture and structural flexibility revealed in the subfamily of large glutamate dehydrogenases by a mycobacterial enzyme.
Commun Biol, 4, 2021
|
|
6Z0F
 
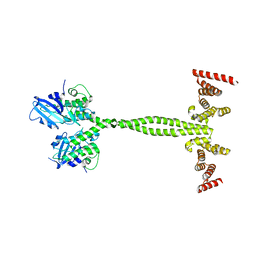 | | Crystal structure of the membrane pseudokinase YukC/EssB from Bacillus subtilis T7SS | | Descriptor: | ESX secretion system protein YukC | | Authors: | Tassinari, M, Bellinzoni, M, Alzari, P.M, Fronzes, R, Gubellini, F. | | Deposit date: | 2020-05-08 | | Release date: | 2021-05-19 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.553 Å) | | Cite: | The Antibacterial Type VII Secretion System of Bacillus subtilis: Structure and Interactions of the Pseudokinase YukC/EssB.
Mbio, 13, 2022
|
|
8P5R
 
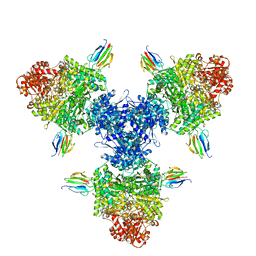 | | Crystal structure of full-length, homohexameric 2-oxoglutarate dehydrogenase KGD from Mycobacterium smegmatis in complex with GarA | | Descriptor: | CALCIUM ION, Glycogen accumulation regulator GarA, MAGNESIUM ION, ... | | Authors: | Wagner, T, Mechaly, A.M, Alzari, P.M, Bellinzoni, M. | | Deposit date: | 2023-05-24 | | Release date: | 2023-08-16 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (4.562 Å) | | Cite: | High resolution cryo-EM and crystallographic snapshots of the actinobacterial two-in-one 2-oxoglutarate dehydrogenase.
Nat Commun, 14, 2023
|
|
1EW3
 
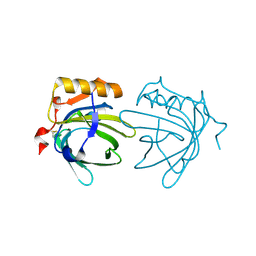 | | CRYSTAL STRUCTURE OF THE MAJOR HORSE ALLERGEN EQU C 1 | | Descriptor: | ALLERGEN EQU C 1 | | Authors: | Lascombe, M.B, Gregoire, C, Poncet, P, Tavares, G.A, Rosinski-Chupin, I, Rabillon, J, Goubran-Botros, H, Mazie, J.C, David, B, Alzari, P.M. | | Deposit date: | 2000-04-21 | | Release date: | 2000-05-03 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of the allergen Equ c 1. A dimeric lipocalin with restricted IgE-reactive epitopes.
J.Biol.Chem., 275, 2000
|
|
4CJ2
 
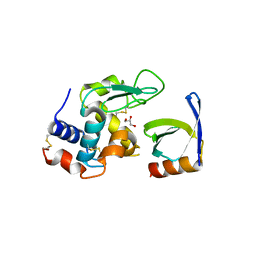 | | Crystal structure of HEWL in complex with affitin H4 | | Descriptor: | AFFITIN H4, GLYCEROL, LYSOZYME C | | Authors: | Correa, A, Pacheco, S, Mechaly, A.E, Obal, G, Behar, G, Mouratou, B, Oppezzo, P, Alzari, P.M, Pecorari, F. | | Deposit date: | 2013-12-18 | | Release date: | 2014-05-21 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Potent and Specific Inhibition of Glycosidases by Small Artificial Binding Proteins (Affitins)
Plos One, 9, 2014
|
|
4CJ0
 
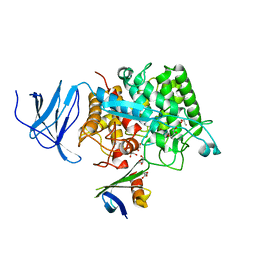 | | Crystal structure of CelD in complex with affitin E12 | | Descriptor: | CALCIUM ION, E12 AFFITIN, ENDOGLUCANASE D, ... | | Authors: | Correa, A, Pacheco, S, Mechaly, A.E, Obal, G, Behar, G, Mouratou, B, Oppezzo, P, Alzari, P.M, Pecorari, F. | | Deposit date: | 2013-12-18 | | Release date: | 2014-05-21 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Potent and Specific Inhibition of Glycosidases by Small Artificial Binding Proteins (Affitins)
Plos One, 9, 2014
|
|
4CJ1
 
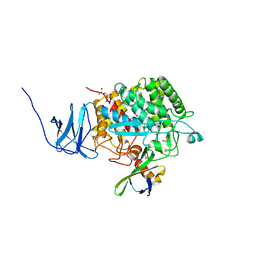 | | Crystal structure of CelD in complex with affitin H3 | | Descriptor: | CALCIUM ION, ENDOGLUCANASE D, GLYCEROL, ... | | Authors: | Correa, A, Pacheco, S, Mechaly, A.E, Obal, G, Behar, G, Mouratou, B, Oppezzo, P, Alzari, P.M, Pecorari, F. | | Deposit date: | 2013-12-18 | | Release date: | 2014-05-21 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Potent and Specific Inhibition of Glycosidases by Small Artificial Binding Proteins (Affitins)
Plos One, 9, 2014
|
|
8BVE
 
 | | MoeA2 from Corynebacterium glutamicum | | Descriptor: | CITRIC ACID, Molybdopterin molybdenumtransferase, SODIUM ION | | Authors: | Martinez, M, Haouz, A, Wehenkel, A.M, Alzari, P.M. | | Deposit date: | 2022-12-03 | | Release date: | 2023-02-22 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | Eukaryotic-like gephyrin and cognate membrane receptor coordinate corynebacterial cell division and polar elongation.
Nat Microbiol, 8, 2023
|
|
8BVF
 
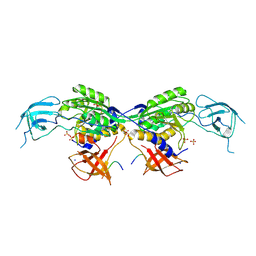 | | MoeA2 from Corynebacterium glutamicum in complex with FtsZ-CTD | | Descriptor: | Cell division protein FtsZ, Molybdopterin molybdenumtransferase, SODIUM ION, ... | | Authors: | Martinez, M, Haouz, A, Wehenkel, A.M, Alzari, P.M. | | Deposit date: | 2022-12-03 | | Release date: | 2023-02-22 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.68 Å) | | Cite: | Eukaryotic-like gephyrin and cognate membrane receptor coordinate corynebacterial cell division and polar elongation.
Nat Microbiol, 8, 2023
|
|
6MFV
 
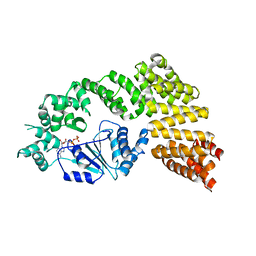 | | Crystal structure of the Signal Transduction ATPase with Numerous Domains (STAND) protein with a tetratricopeptide repeat sensor PH0952 from Pyrococcus horikoshii | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, tetratricopeptide repeat sensor PH0952 | | Authors: | Lisa, M.N, Alzari, P.M, Haouz, A, Danot, O. | | Deposit date: | 2018-09-12 | | Release date: | 2019-02-20 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Double autoinhibition mechanism of signal transduction ATPases with numerous domains (STAND) with a tetratricopeptide repeat sensor.
Nucleic Acids Res., 47, 2019
|
|
7JSR
 
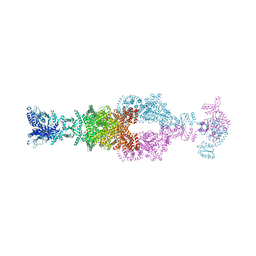 | | Crystal structure of the large glutamate dehydrogenase composed of 180 kDa subunits from Mycobacterium smegmatis | | Descriptor: | NAD-specific glutamate dehydrogenase | | Authors: | Lazaro, M, Melero, R, Huet, C, Lopez-Alonso, J.P, Delgado, S, Dodu, A, Bruch, E.M, Abriata, L.A, Alzari, P.M, Valle, M, Lisa, M.N. | | Deposit date: | 2020-08-15 | | Release date: | 2021-06-09 | | Last modified: | 2025-05-14 | | Method: | X-RAY DIFFRACTION (6.27 Å) | | Cite: | 3D architecture and structural flexibility revealed in the subfamily of large glutamate dehydrogenases by a mycobacterial enzyme.
Commun Biol, 4, 2021
|
|
2FUM
 
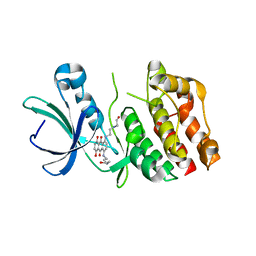 | | Catalytic domain of protein kinase PknB from Mycobacterium tuberculosis in complex with mitoxantrone | | Descriptor: | 1,4-DIHYDROXY-5,8-BIS({2-[(2-HYDROXYETHYL)AMINO]ETHYL}AMINO)-9,10-ANTHRACENEDIONE, Probable serine/threonine-protein kinase pknB | | Authors: | Wehenkel, A, Alzari, P.M. | | Deposit date: | 2006-01-27 | | Release date: | 2006-08-01 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.89 Å) | | Cite: | The structure of PknB in complex with mitoxantrone, an ATP-competitive inhibitor, suggests a mode of protein kinase regulation in mycobacteria
Febs Lett., 580, 2006
|
|
