6WLH
 
 | |
1F68
 
 | |
9PCY
 
 | | HIGH-RESOLUTION SOLUTION STRUCTURE OF REDUCED FRENCH BEAN PLASTOCYANIN AND COMPARISON WITH THE CRYSTAL STRUCTURE OF POPLAR PLASTOCYANIN | | Descriptor: | COPPER (II) ION, PLASTOCYANIN | | Authors: | Moore, J.M, Lepre, C.A, Gippert, G.P, Chazin, W.J, Case, D.A, Wright, P.E. | | Deposit date: | 1991-03-18 | | Release date: | 1993-10-31 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | High-resolution solution structure of reduced French bean plastocyanin and comparison with the crystal structure of poplar plastocyanin.
J.Mol.Biol., 221, 1991
|
|
8W2W
 
 | | Structure of transthyretin synthetic mutation A120L | | Descriptor: | Transthyretin | | Authors: | Yang, K, Sun, X, Ferguson, J.A, Stanfield, R.L, Wright, P.E. | | Deposit date: | 2024-02-21 | | Release date: | 2024-02-28 | | Last modified: | 2024-08-28 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Mispacking of the F87 sidechain drives aggregation-promoting conformational fluctuations in the subunit interfaces of the transthyretin tetramer.
Protein Sci., 33, 2024
|
|
8W1N
 
 | |
2KJE
 
 | | NMR structure of CBP TAZ2 and adenoviral E1A complex | | Descriptor: | CREB-binding protein, Early E1A 32 kDa protein, ZINC ION | | Authors: | Ferreon, J.C, Martinez-Yamout, M, Dyson, H, Wright, P.E. | | Deposit date: | 2009-05-27 | | Release date: | 2009-09-15 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structural basis for subversion of cellular control mechanisms by the adenoviral E1A oncoprotein.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
2L14
 
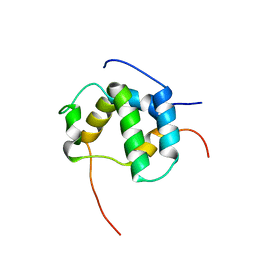 | | Structure of CBP nuclear coactivator binding domain in complex with p53 TAD | | Descriptor: | CREB-binding protein, Cellular tumor antigen p53 | | Authors: | Lee, C, Martinez-Yamout, M.A, Dyson, H.J, Wright, P.E. | | Deposit date: | 2010-07-22 | | Release date: | 2010-11-03 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structure of the p53 transactivation domain in complex with the nuclear receptor coactivator binding domain of CREB binding protein.
Biochemistry, 49, 2010
|
|
2LT7
 
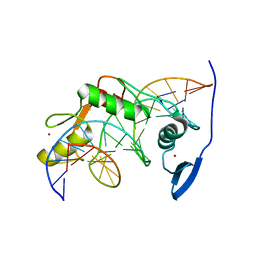 | | Solution NMR structure of Kaiso zinc finger DNA binding domain in complex with Kaiso binding site DNA | | Descriptor: | DNA (5'-D(*CP*GP*TP*TP*AP*TP*TP*GP*GP*CP*AP*GP*GP*AP*AP*GP*CP*AP*C)-3'), DNA (5'-D(*GP*TP*GP*CP*TP*TP*CP*CP*TP*GP*CP*CP*AP*AP*TP*AP*AP*CP*G)-3'), Transcriptional regulator Kaiso, ... | | Authors: | Buck-Koehntop, B.A, Stanfield, R.L, Ekiert, D.C, Martinez-Yamout, M.A, Dyson, H, Wilson, I.A, Wright, P.E. | | Deposit date: | 2012-05-15 | | Release date: | 2012-09-05 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Molecular basis for recognition of methylated and specific DNA sequences by the zinc finger protein Kaiso.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
1XOB
 
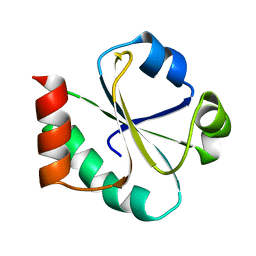 | | THIOREDOXIN (REDUCED DITHIO FORM), NMR, 20 STRUCTURES | | Descriptor: | THIOREDOXIN | | Authors: | Jeng, M.-F, Campbell, A.P, Begley, T, Holmgren, A, Case, D.A, Wright, P.E, Dyson, H.J. | | Deposit date: | 1995-11-28 | | Release date: | 1996-06-10 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | High-resolution solution structures of oxidized and reduced Escherichia coli thioredoxin.
Structure, 2, 1994
|
|
1XOA
 
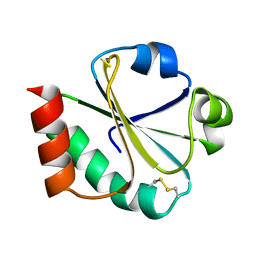 | | THIOREDOXIN (OXIDIZED DISULFIDE FORM), NMR, 20 STRUCTURES | | Descriptor: | THIOREDOXIN | | Authors: | Jeng, M.-F, Campbell, A.P, Begley, T, Holmgren, A, Case, D.A, Wright, P.E, Dyson, H.J. | | Deposit date: | 1995-11-28 | | Release date: | 1996-06-10 | | Last modified: | 2017-11-29 | | Method: | SOLUTION NMR | | Cite: | High-resolution solution structures of oxidized and reduced Escherichia coli thioredoxin.
Structure, 2, 1994
|
|
1ZNF
 
 | | THREE-DIMENSIONAL SOLUTION STRUCTURE OF A SINGLE ZINC FINGER DNA-BINDING DOMAIN | | Descriptor: | 31ST ZINC FINGER FROM XFIN, ZINC ION | | Authors: | Lee, M.S, Gippert, G.P, Soman, K.V, Case, D.A, Wright, P.E. | | Deposit date: | 1989-09-25 | | Release date: | 1991-07-15 | | Last modified: | 2017-11-29 | | Method: | SOLUTION NMR | | Cite: | Three-dimensional solution structure of a single zinc finger DNA-binding domain.
Science, 245, 1989
|
|
1AX3
 
 | | SOLUTION NMR STRUCTURE OF B. SUBTILIS IIAGLC, 16 STRUCTURES | | Descriptor: | GLUCOSE PERMEASE IIA DOMAIN | | Authors: | Chen, Y, Case, D.A, Reizer, J, Saier Junior, M.H, Wright, P.E. | | Deposit date: | 1997-10-25 | | Release date: | 1998-06-17 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | High-resolution solution structure of Bacillus subtilis IIAglc.
Proteins, 31, 1998
|
|
1MYF
 
 | | SOLUTION STRUCTURE OF CARBONMONOXY MYOGLOBIN DETERMINED FROM NMR DISTANCE AND CHEMICAL SHIFT CONSTRAINTS | | Descriptor: | CARBON MONOXIDE, MYOGLOBIN, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Osapay, K, Theriault, Y, Wright, P.E, Case, D.A. | | Deposit date: | 1994-12-02 | | Release date: | 1995-02-27 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of carbonmonoxy myoglobin determined from nuclear magnetic resonance distance and chemical shift constraints.
J.Mol.Biol., 244, 1994
|
|
8T5X
 
 | | Probing the dissociation pathway of a kinetically labile transthyretin mutant (A25T) | | Descriptor: | Transthyretin | | Authors: | Ferguson, J.A, Sun, X, Leach, B.I, Stanfield, R.L, Dyson, H.J, Wright, P.E. | | Deposit date: | 2023-06-14 | | Release date: | 2023-08-02 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Probing the Dissociation Pathway of a Kinetically Labile Transthyretin Mutant.
J.Am.Chem.Soc., 146, 2024
|
|
1TF3
 
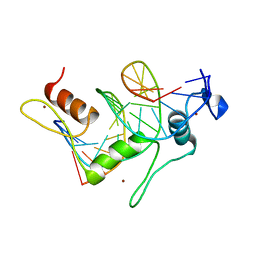 | | TFIIIA FINGER 1-3 BOUND TO DNA, NMR, 22 STRUCTURES | | Descriptor: | 5S RNA GENE, TRANSCRIPTION FACTOR IIIA, ZINC ION | | Authors: | Foster, M.P, Wuttke, D.S, Radhakrishnan, I, Case, D.A, Gottesfeld, J.M, Wright, P.E. | | Deposit date: | 1997-07-01 | | Release date: | 1997-09-17 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Domain packing and dynamics in the DNA complex of the N-terminal zinc fingers of TFIIIA.
Nat.Struct.Biol., 4, 1997
|
|
3QL3
 
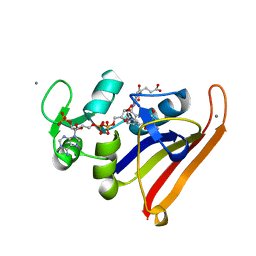 | | Re-refined coordinates for PDB entry 1RX2 | | Descriptor: | Dihydrofolate reductase, FOLIC ACID, MANGANESE (II) ION, ... | | Authors: | Bhabha, G, Ekiert, D.C, Wright, P.E, Wilson, I.A. | | Deposit date: | 2011-02-02 | | Release date: | 2011-04-27 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | A dynamic knockout reveals that conformational fluctuations influence the chemical step of enzyme catalysis.
Science, 332, 2011
|
|
3QL0
 
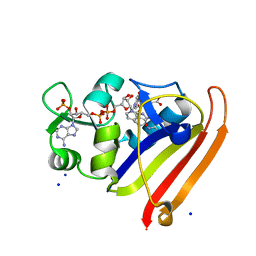 | | Crystal structure of N23PP/S148A mutant of E. coli dihydrofolate reductase | | Descriptor: | Dihydrofolate reductase, FOLIC ACID, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Bhabha, G, Ekiert, D.C, Wright, P.E, Wilson, I.A. | | Deposit date: | 2011-02-02 | | Release date: | 2011-04-20 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | A dynamic knockout reveals that conformational fluctuations influence the chemical step of enzyme catalysis.
Science, 332, 2011
|
|
4F6N
 
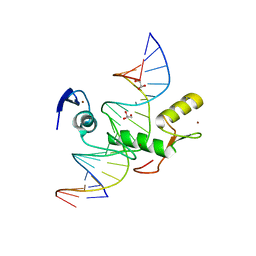 | | Crystal structure of Kaiso zinc finger DNA binding protein in complex with methylated CpG site DNA | | Descriptor: | DNA (5'-D(*CP*GP*TP*AP*TP*AP*GP*AP*(5CM)P*GP*(5CM)P*GP*GP*TP*GP*AP*CP*AP*C)-3'), DNA (5'-D(*GP*TP*GP*TP*CP*AP*CP*(5CM)P*GP*(5CM)P*GP*TP*CP*TP*AP*TP*AP*CP*G)-3'), GLYCEROL, ... | | Authors: | Buck-Koehntop, B.A, Stanfield, R.L, Ekiert, D.C, Martinez-Yamout, M.A, Dyson, H.J, Wilson, I.A, Wright, P.E. | | Deposit date: | 2012-05-15 | | Release date: | 2012-09-05 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Molecular basis for recognition of methylated and specific DNA sequences by the zinc finger protein Kaiso.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
4F6M
 
 | | Crystal structure of Kaiso zinc finger DNA binding domain in complex with Kaiso binding site DNA | | Descriptor: | DNA (5'-D(*CP*GP*TP*TP*AP*TP*TP*GP*GP*CP*AP*GP*GP*AP*AP*GP*CP*AP*C)-3'), DNA (5'-D(*GP*TP*GP*CP*TP*TP*CP*CP*TP*GP*CP*CP*AP*AP*TP*AP*AP*CP*G)-3'), Transcriptional regulator Kaiso, ... | | Authors: | Buck-Koehntop, B.A, Stanfield, R.L, Ekiert, D.C, Martinez-Yamout, M.A, Dyson, H.J, Wilson, I.A, Wright, P.E. | | Deposit date: | 2012-05-15 | | Release date: | 2012-09-05 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Molecular basis for recognition of methylated and specific DNA sequences by the zinc finger protein Kaiso.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
5HPD
 
 | | Solution Structure of TAZ2-p53TAD | | Descriptor: | CREB-binding protein,Cellular tumor antigen p53 fusion protein, ZINC ION | | Authors: | Krois, A.S, Ferreon, J.C, Martinez-Yamout, M.A, Dyson, H.J, Wright, P.E. | | Deposit date: | 2016-01-20 | | Release date: | 2016-03-16 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Recognition of the disordered p53 transactivation domain by the transcriptional adapter zinc finger domains of CREB-binding protein.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
5HOU
 
 | | Solution Structure of p53TAD-TAZ1 | | Descriptor: | Cellular tumor antigen p53,CREB-binding protein fusion protein, ZINC ION | | Authors: | Krois, A.S, Ferreon, J.C, Martinez-Yamout, M.A, Dyson, H.J, Wright, P.E. | | Deposit date: | 2016-01-19 | | Release date: | 2016-03-16 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Recognition of the disordered p53 transactivation domain by the transcriptional adapter zinc finger domains of CREB-binding protein.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
4KJJ
 
 | | Cryogenic WT DHFR | | Descriptor: | Dihydrofolate reductase, FOLIC ACID, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | van den Bedem, H, Bhabha, G, Yang, K, Wright, P.E, Fraser, J.S. | | Deposit date: | 2013-05-03 | | Release date: | 2013-08-21 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Automated identification of functional dynamic contact networks from X-ray crystallography.
Nat.Methods, 10, 2013
|
|
4KJL
 
 | | Room Temperature N23PPS148A DHFR | | Descriptor: | Dihydrofolate reductase, FOLIC ACID, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | van den Bedem, H, Bhabha, G, Yang, K, Wright, P.E, Fraser, J.S. | | Deposit date: | 2013-05-03 | | Release date: | 2013-08-21 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.38 Å) | | Cite: | Automated identification of functional dynamic contact networks from X-ray crystallography.
Nat.Methods, 10, 2013
|
|
5HP0
 
 | | Solution Structure of TAZ2-p53AD2 | | Descriptor: | CREB-binding protein,Cellular tumor antigen p53 fusion protein, ZINC ION | | Authors: | Krois, A.S, Ferreon, J.C, Martinez-Yamout, M.A, Dyson, H.J, Wright, P.E. | | Deposit date: | 2016-01-19 | | Release date: | 2016-03-16 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Recognition of the disordered p53 transactivation domain by the transcriptional adapter zinc finger domains of CREB-binding protein.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
4KJK
 
 | | Room Temperature WT DHFR | | Descriptor: | CALCIUM ION, Dihydrofolate reductase, FOLIC ACID, ... | | Authors: | van den Bedem, H, Bhabha, G, Yang, K, Wright, P.E, Fraser, J.S. | | Deposit date: | 2013-05-03 | | Release date: | 2013-08-21 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.351 Å) | | Cite: | Automated identification of functional dynamic contact networks from X-ray crystallography.
Nat.Methods, 10, 2013
|
|
