6XFR
 
 | |
4KH3
 
 | | Structure of a bacterial self-associating protein | | Descriptor: | Antigen 43, MALONATE ION | | Authors: | Heras, B, Gee, C.L, Schembri, M.A, Totsika, M. | | Deposit date: | 2013-04-30 | | Release date: | 2014-01-15 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The antigen 43 structure reveals a molecular Velcro-like mechanism of autotransporter-mediated bacterial clumping.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
5EK5
 
 | | STRUCTURAL CHARACTERIZATION OF IRMA FROM ESCHERICHIA COLI | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, FORMIC ACID, ... | | Authors: | Heras, B, Moriel, D.G, Paxman, J.J, Schembri, M.A. | | Deposit date: | 2015-11-03 | | Release date: | 2016-03-09 | | Last modified: | 2016-07-13 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | Molecular and Structural Characterization of a Novel Escherichia coli Interleukin Receptor Mimic Protein.
Mbio, 7, 2016
|
|
4C7M
 
 | | The crystal structure of TcpB or BtpA TIR domain | | Descriptor: | Toll/interleukin-1 receptor domain-containing protein | | Authors: | Alaidarous, M, Ve, T, Casey, L.W, Valkov, E, Ullah, M.O, Schembri, M.A, Mansell, A, Sweet, M.J, Kobe, B. | | Deposit date: | 2013-09-23 | | Release date: | 2013-12-04 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.57 Å) | | Cite: | Mechanism of bacterial interference with TLR4 signaling by Brucella Toll/interleukin-1 receptor domain-containing protein TcpB.
J.Biol.Chem., 289, 2014
|
|
6N8A
 
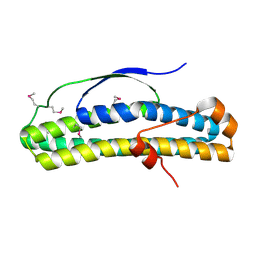 | | Crystal structure of selenomethionine-containing AcaB from uropathogenic E. coli | | Descriptor: | CHLORIDE ION, transcription regulator AcaB | | Authors: | Luo, Z, Hancock, S.J, Schembri, M.A, Kobe, B. | | Deposit date: | 2018-11-28 | | Release date: | 2020-07-15 | | Last modified: | 2021-01-27 | | Method: | X-RAY DIFFRACTION (3.4011 Å) | | Cite: | Comprehensive analysis of IncC plasmid conjugation identifies a crucial role for the transcriptional regulator AcaB.
Nat Microbiol, 5, 2020
|
|
6N8B
 
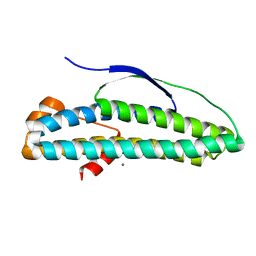 | | Crystal structure of transcription regulator AcaB from uropathogenic E. coli | | Descriptor: | CALCIUM ION, transcription regulator AcaB | | Authors: | Luo, Z, Hancock, S.J, Schembri, M.A, Kobe, B. | | Deposit date: | 2018-11-29 | | Release date: | 2020-07-15 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.94 Å) | | Cite: | Comprehensive analysis of IncC plasmid conjugation identifies a crucial role for the transcriptional regulator AcaB.
Nat Microbiol, 5, 2020
|
|
4GXZ
 
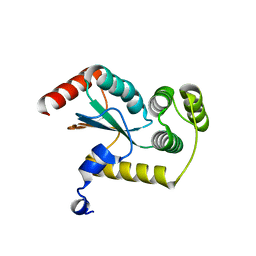 | | Crystal structure of a periplasmic thioredoxin-like protein from Salmonella enterica serovar Typhimurium | | Descriptor: | Suppression of copper sensitivity protein | | Authors: | Shepherd, M, Heras, B, King, G.J, Argente, M.P, Achard, M.E.S, King, N.P, McEwan, A.G, Schembri, M.A. | | Deposit date: | 2012-09-04 | | Release date: | 2013-07-17 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Structural and functional characterization of ScsC, a periplasmic thioredoxin-like protein from Salmonella enterica serovar Typhimurium
Antioxid Redox Signal, 19, 2013
|
|
6AQJ
 
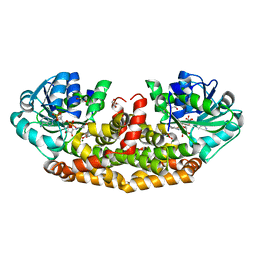 | | Crystal structures of Staphylococcus aureus ketol-acid reductoisomerase in complex with two transition state analogs that have biocidal activity. | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, Ketol-acid reductoisomerase (NADP(+)), ... | | Authors: | Patel, K.M, Teran, D, Zheng, S, Gracia, M, Lv, Y, Schembri, M.A, McGeary, R.P, Schenk, G, Guddat, L.W. | | Deposit date: | 2017-08-20 | | Release date: | 2017-10-18 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.373 Å) | | Cite: | Crystal Structures of Staphylococcus aureus Ketol-Acid Reductoisomerase in Complex with Two Transition State Analogues that Have Biocidal Activity.
Chemistry, 23, 2017
|
|
6BUL
 
 | | Crystal structure of Staphylococcus aureus ketol-acid reductoisomerase with hydroxyoxamate inhibitor 2 | | Descriptor: | Ketol-acid reductoisomerase (NADP(+)), MAGNESIUM ION, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Kandale, A, Patel, K.M, Zheng, S, You, L, Guddat, L.W, Schenk, G, Schembri, M.A, McFeary, R.P. | | Deposit date: | 2017-12-10 | | Release date: | 2018-12-12 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Design, synthesis, in vitro activity and crystallisation of novel N-isopropyl-N-hydroxyoxamate derivatives as ketol-acid reductoisomerase (KARI) inhibitors
To Be Published
|
|
6C5N
 
 | | Crystal structure of Staphylococcus aureus ketol-acid reductoisomerase with hydroxyoxamate inhibitor 1 | | Descriptor: | (cyclopentylamino)(oxo)acetic acid, IMIDAZOLE, Ketol-acid reductoisomerase (NADP(+)), ... | | Authors: | Kandale, A, Patel, K.M, Zheng, S, You, L, Guddat, L.W, Schenk, G, Schembri, M.A, McGeary, R.P. | | Deposit date: | 2018-01-16 | | Release date: | 2019-01-30 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.673 Å) | | Cite: | Design, synthesis, in vitro activity and crystallisation of novel N-isopropyl-N-hydroxyoxamate derivatives as ketol-acid reductosiomerase (KARI) inhibitor
To Be Published
|
|
7MZQ
 
 | | Crystal structure of the UcaD lectin-binding domain in complex with fucose | | Descriptor: | CHLORIDE ION, Fimbrial adhesin UcaD, beta-L-fucopyranose | | Authors: | Ve, T, Lo, A.W, Schembri, M.A, Kobe, B. | | Deposit date: | 2021-05-24 | | Release date: | 2022-07-06 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Ucl fimbriae regulation and glycan receptor specificity contribute to gut colonisation by extra-intestinal pathogenic Escherichia coli.
Plos Pathog., 18, 2022
|
|
7MZO
 
 | | Crystal structure of the UcaD lectin-binding domain | | Descriptor: | CHLORIDE ION, Fimbrial adhesin UcaD | | Authors: | Ve, T, Lo, A.W, Schembri, M.A, Kobe, B. | | Deposit date: | 2021-05-24 | | Release date: | 2022-07-06 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Ucl fimbriae regulation and glycan receptor specificity contribute to gut colonisation by extra-intestinal pathogenic Escherichia coli.
Plos Pathog., 18, 2022
|
|
7MZS
 
 | | Crystal structure of the UcaD lectin-binding domain in complex with galactose | | Descriptor: | CHLORIDE ION, Fimbrial adhesin UcaD, alpha-D-galactopyranose | | Authors: | Ve, T, Lo, A.W, Schembri, M.A, Kobe, B. | | Deposit date: | 2021-05-24 | | Release date: | 2022-07-06 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Ucl fimbriae regulation and glycan receptor specificity contribute to gut colonisation by extra-intestinal pathogenic Escherichia coli.
Plos Pathog., 18, 2022
|
|
7MZP
 
 | | Crystal structure of the UclD lectin-binding domain | | Descriptor: | F17-like fimbril adhesin subunit UclD, IODIDE ION | | Authors: | Ve, T, Lo, A.W, Schembri, M.A, Kobe, B. | | Deposit date: | 2021-05-24 | | Release date: | 2022-07-06 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Ucl fimbriae regulation and glycan receptor specificity contribute to gut colonisation by extra-intestinal pathogenic Escherichia coli.
Plos Pathog., 18, 2022
|
|
7MZR
 
 | | Crystal structure of the UcaD lectin-binding domain in complex with glucose | | Descriptor: | CHLORIDE ION, Fimbrial adhesin UcaD, beta-D-glucopyranose | | Authors: | Ve, T, Lo, A.W, Schembri, M.A, Kobe, B. | | Deposit date: | 2021-05-24 | | Release date: | 2022-07-06 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Ucl fimbriae regulation and glycan receptor specificity contribute to gut colonisation by extra-intestinal pathogenic Escherichia coli.
Plos Pathog., 18, 2022
|
|
3L9S
 
 | |
5W3K
 
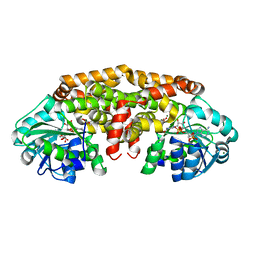 | | Crystal structure of Staphylococcus aureus ketol-acid reductoisomerase in complex NADPH, Mg2+ and CPD | | Descriptor: | Ketol-acid reductoisomerase (NADP(+)), MAGNESIUM ION, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Patel, K.M, Teran, D, Zheng, S, Kandale, A, Schembri, M, McGeary, R.P, Schenk, G, Guddat, L.W. | | Deposit date: | 2017-06-08 | | Release date: | 2017-10-25 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.589 Å) | | Cite: | Crystal Structures of Staphylococcus aureus Ketol-Acid Reductoisomerase in Complex with Two Transition State Analogues that Have Biocidal Activity.
Chemistry, 23, 2017
|
|
5ID4
 
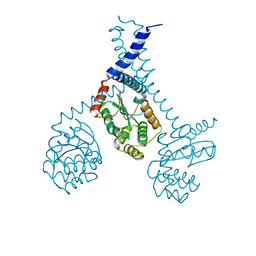 | |
5IDR
 
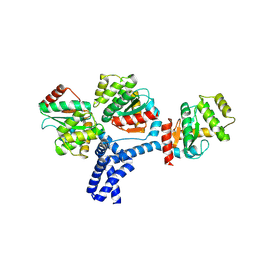 | |
4XVW
 
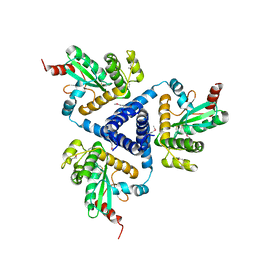 | |
7UXU
 
 | | CryoEM structure of the TIR domain from AbTir in complex with 3AD | | Descriptor: | Molecular chaperone Tir, [[(2~{R},3~{S},4~{R},5~{R})-5-(6-aminopurin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methoxy-oxidanyl-phosphoryl] [(2~{R},3~{S},4~{R},5~{R})-5-(8-azanylisoquinolin-2-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methyl hydrogen phosphate | | Authors: | Li, S, Nanson, J.D, Manik, M.K, Gu, W, Landsberg, M.J, Ve, T, Kobe, B. | | Deposit date: | 2022-05-06 | | Release date: | 2022-09-07 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (2.74 Å) | | Cite: | Cyclic ADP ribose isomers: Production, chemical structures, and immune signaling.
Science, 377, 2022
|
|
7UXS
 
 | | Crystal structure of the BcThsA SLOG domain in complex with 3'cADPR | | Descriptor: | (2R,3R,3aS,5S,6R,7S,8R,11R,13S,15aR)-2-(6-amino-9H-purin-9-yl)-3,6,7,11,13-pentahydroxyoctahydro-2H,5H,11H,13H-5,8-epoxy-11lambda~5~,13lambda~5~-furo[2,3-g][1,3,5,9,2,4]tetraoxadiphosphacyclotetradecine-11,13-dione, BcThsA, GLYCEROL, ... | | Authors: | Shi, Y, Masic, V, Mosaiab, T, Ve, T. | | Deposit date: | 2022-05-06 | | Release date: | 2022-09-07 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | Cyclic ADP ribose isomers: Production, chemical structures, and immune signaling.
Science, 377, 2022
|
|
7UXT
 
 | | Crystal structure of ligand-free SeThsA | | Descriptor: | GLYCEROL, TRIETHYLENE GLYCOL, USG protein | | Authors: | Shi, Y, Masic, V, Mosaiab, T, Nanson, J.D, Kobe, B, Ve, T. | | Deposit date: | 2022-05-06 | | Release date: | 2022-09-07 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Cyclic ADP ribose isomers: Production, chemical structures, and immune signaling.
Science, 377, 2022
|
|
7UWG
 
 | | The crystal structure of the TIR domain-containing protein from Acinetobacter baumannii (AbTir) | | Descriptor: | HEXAETHYLENE GLYCOL, Molecular chaperone Tir, SULFATE ION | | Authors: | Manik, M.K, Nanson, J.D, Ve, T, Kobe, B. | | Deposit date: | 2022-05-03 | | Release date: | 2022-09-07 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.16 Å) | | Cite: | Cyclic ADP ribose isomers: Production, chemical structures, and immune signaling.
Science, 377, 2022
|
|
7UXR
 
 | | Crystal structure of the BtTir TIR domain | | Descriptor: | TIR domain protein | | Authors: | Shi, Y, Masic, V, Mosaiab, T, Vasquez, E, Ve, T. | | Deposit date: | 2022-05-06 | | Release date: | 2022-09-07 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.42 Å) | | Cite: | Cyclic ADP ribose isomers: Production, chemical structures, and immune signaling.
Science, 377, 2022
|
|
