6V00
 
 | | structure of human KCNQ1-KCNE3-CaM complex | | Descriptor: | CALCIUM ION, Calmodulin-1, MCherry fluorescent protein,Potassium voltage-gated channel subfamily E member 3, ... | | Authors: | Mackinnon, R, Sun, J. | | Deposit date: | 2019-11-16 | | Release date: | 2019-12-04 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural Basis of Human KCNQ1 Modulation and Gating.
Cell, 180, 2020
|
|
6UZZ
 
 | | structure of human KCNQ1-CaM complex | | Descriptor: | CALCIUM ION, Calmodulin-1, Potassium voltage-gated channel subfamily KQT member 1 | | Authors: | Mackinnon, R, Sun, J. | | Deposit date: | 2019-11-16 | | Release date: | 2019-12-04 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural Basis of Human KCNQ1 Modulation and Gating.
Cell, 180, 2020
|
|
6V01
 
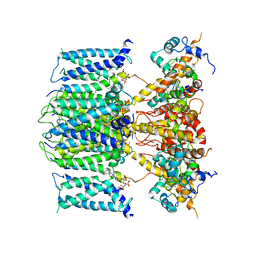 | | structure of human KCNQ1-KCNE3-CaM complex with PIP2 | | Descriptor: | CALCIUM ION, Calmodulin-1, Potassium voltage-gated channel subfamily E member 3, ... | | Authors: | Mackinnon, R, Sun, J. | | Deposit date: | 2019-11-18 | | Release date: | 2019-12-04 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Structural Basis of Human KCNQ1 Modulation and Gating.
Cell, 180, 2020
|
|
4JTA
 
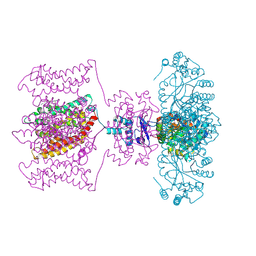 | | Crystal structure of Kv1.2-2.1 paddle chimera channel in complex with Charybdotoxin | | Descriptor: | (1R)-2-{[(S)-{[(2S)-2,3-dihydroxypropyl]oxy}(hydroxy)phosphoryl]oxy}-1-[(hexadecanoyloxy)methyl]ethyl (9Z)-octadec-9-enoate, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, POTASSIUM ION, ... | | Authors: | MacKinnon, R, Banerjee, A, Lee, A, Campbell, E. | | Deposit date: | 2013-03-23 | | Release date: | 2013-06-12 | | Last modified: | 2019-12-25 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure of a pore-blocking toxin in complex with a eukaryotic voltage-dependent K(+) channel.
Elife, 2, 2013
|
|
5TJ6
 
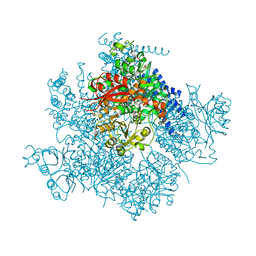 | | Ca2+ bound aplysia Slo1 | | Descriptor: | (1R)-2-{[(S)-{[(2S)-2,3-dihydroxypropyl]oxy}(hydroxy)phosphoryl]oxy}-1-[(hexadecanoyloxy)methyl]ethyl (9Z)-octadec-9-enoate, CALCIUM ION, High conductance calcium-activated potassium channel, ... | | Authors: | MacKinnon, R, Tao, X, Hite, R.K. | | Deposit date: | 2016-10-03 | | Release date: | 2016-12-14 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Cryo-EM structure of the open high-conductance Ca(2+)-activated K(+) channel.
Nature, 541, 2017
|
|
5TJI
 
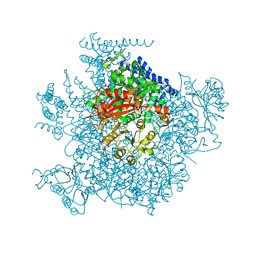 | | Ca2+ bound aplysia Slo1 | | Descriptor: | (1R)-2-{[(S)-{[(2S)-2,3-dihydroxypropyl]oxy}(hydroxy)phosphoryl]oxy}-1-[(hexadecanoyloxy)methyl]ethyl (9Z)-octadec-9-enoate, High conductance calcium-activated potassium channel | | Authors: | MacKinnon, R, Tao, X, Hite, R.K. | | Deposit date: | 2016-10-04 | | Release date: | 2016-12-28 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Structural basis for gating the high-conductance Ca(2+)-activated K(+) channel.
Nature, 541, 2017
|
|
5VMS
 
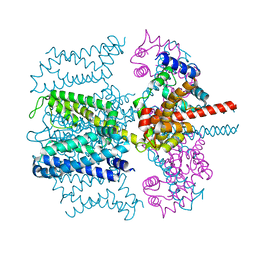 | | CryoEM structure of Xenopus KCNQ1 channel | | Descriptor: | CALCIUM ION, Calmodulin, Potassium voltage-gated channel subfamily KQT member 1 | | Authors: | Mackinnon, R, Sun, J. | | Deposit date: | 2017-04-28 | | Release date: | 2017-06-07 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Cryo-EM Structure of a KCNQ1/CaM Complex Reveals Insights into Congenital Long QT Syndrome.
Cell, 169, 2017
|
|
1S5H
 
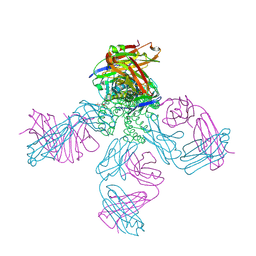 | | Potassium Channel Kcsa-Fab Complex T75C mutant in K+ | | Descriptor: | ANTIBODY FAB FRAGMENT HEAVY CHAIN, ANTIBODY FAB FRAGMENT LIGHT CHAIN, DIACYL GLYCEROL, ... | | Authors: | Mackinnon, R, Zhou, M. | | Deposit date: | 2004-01-20 | | Release date: | 2004-05-18 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | A mutant KcsA K(+) channel with altered conduction properties and selectivity filter ion distribution.
J.Mol.Biol., 338, 2004
|
|
1AGT
 
 | | SOLUTION STRUCTURE OF THE POTASSIUM CHANNEL INHIBITOR AGITOXIN 2: CALIPER FOR PROBING CHANNEL GEOMETRY | | Descriptor: | AGITOXIN 2 | | Authors: | Krezel, A.M, Kasibhatla, C, Hidalgo, P, Mackinnon, R, Wagner, G. | | Deposit date: | 1995-04-14 | | Release date: | 1995-07-10 | | Last modified: | 2017-11-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the potassium channel inhibitor agitoxin 2: caliper for probing channel geometry.
Protein Sci., 4, 1995
|
|
8UQN
 
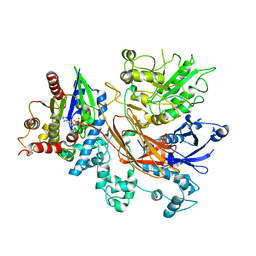 | | PLCb3-Gaq complex on membranes | | Descriptor: | 1-phosphatidylinositol 4,5-bisphosphate phosphodiesterase beta-3, CALCIUM ION, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Falzone, M.E, MacKinnon, R. | | Deposit date: | 2023-10-24 | | Release date: | 2023-12-06 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | The mechanism of G alpha q regulation of PLC beta 3 -catalyzed PIP2 hydrolysis.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
8UQO
 
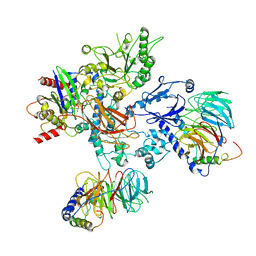 | | PLCb3-Gbg-Gaq complex on membranes | | Descriptor: | 1-phosphatidylinositol 4,5-bisphosphate phosphodiesterase beta-3, CALCIUM ION, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Falzone, M.E, MacKinnon, R. | | Deposit date: | 2023-10-24 | | Release date: | 2023-12-06 | | Method: | ELECTRON MICROSCOPY (3.37 Å) | | Cite: | The mechanism of G alpha q regulation of PLC beta 3 -catalyzed PIP2 hydrolysis.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
5K7L
 
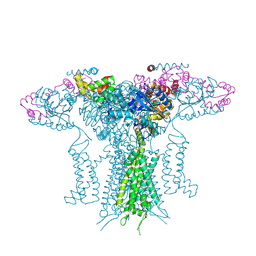 | |
4WFH
 
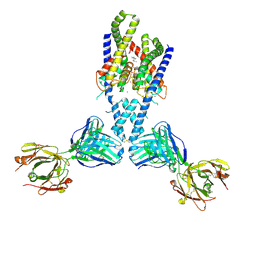 | |
4WFG
 
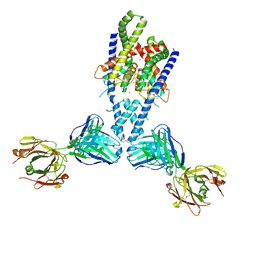 | | Human TRAAK K+ channel in a Tl+ bound conductive conformation | | Descriptor: | ANTI-TRAAK ANTIBODY 13E9 FAB FRAGMENT HEAVY CHAIN, ANTI-TRAAK ANTIBODY 13E9 FAB FRAGMENT LIGHT CHAIN, CALCIUM ION, ... | | Authors: | Brohawn, S.G, MacKinnon, R. | | Deposit date: | 2014-09-15 | | Release date: | 2014-12-03 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Physical mechanism for gating and mechanosensitivity of the human TRAAK K+ channel.
Nature, 516, 2014
|
|
4WFE
 
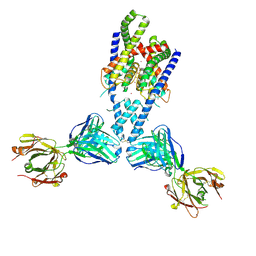 | | Human TRAAK K+ channel in a K+ bound conductive conformation | | Descriptor: | ANTI-TRAAK ANTIBODY 13E9 FAB FRAGMENT HEAVY CHAIN, ANTI-TRAAK ANTIBODY 13E9 FAB FRAGMENT LIGHT CHAIN, CALCIUM ION, ... | | Authors: | Brohawn, S.G, MacKinnon, R. | | Deposit date: | 2014-09-15 | | Release date: | 2014-12-03 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Physical mechanism for gating and mechanosensitivity of the human TRAAK K+ channel.
Nature, 516, 2014
|
|
4WFF
 
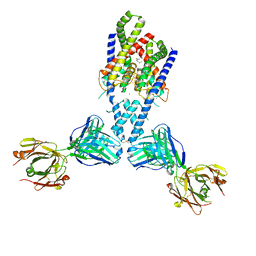 | |
1BFE
 
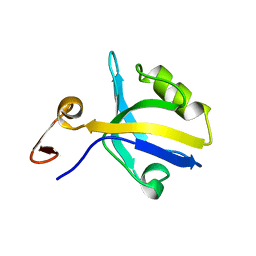 | | THE THIRD PDZ DOMAIN FROM THE SYNAPTIC PROTEIN PSD-95 | | Descriptor: | PSD-95 | | Authors: | Doyle, D.A, Lee, A, Lewis, J, Kim, E, Sheng, M, Mackinnon, R. | | Deposit date: | 1998-05-20 | | Release date: | 1998-10-21 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structures of a complexed and peptide-free membrane protein-binding domain: molecular basis of peptide recognition by PDZ.
Cell(Cambridge,Mass.), 85, 1996
|
|
1BE9
 
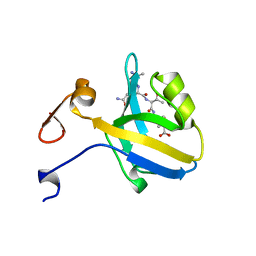 | | THE THIRD PDZ DOMAIN FROM THE SYNAPTIC PROTEIN PSD-95 IN COMPLEX WITH A C-TERMINAL PEPTIDE DERIVED FROM CRIPT. | | Descriptor: | CRIPT, PSD-95 | | Authors: | Doyle, D.A, Lee, A, Lewis, J, Kim, E, Sheng, M, Mackinnon, R. | | Deposit date: | 1998-05-20 | | Release date: | 1998-10-21 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Crystal structures of a complexed and peptide-free membrane protein-binding domain: molecular basis of peptide recognition by PDZ.
Cell(Cambridge,Mass.), 85, 1996
|
|
1BL8
 
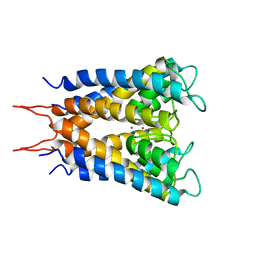 | | POTASSIUM CHANNEL (KCSA) FROM STREPTOMYCES LIVIDANS | | Descriptor: | POTASSIUM ION, PROTEIN (POTASSIUM CHANNEL PROTEIN) | | Authors: | Doyle, D.A, Cabral, J.M, Pfuetzner, R.A, Kuo, A, Gulbis, J.M, Cohen, S.L, Chait, B.T, Mackinnon, R. | | Deposit date: | 1998-07-23 | | Release date: | 1998-07-29 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | The structure of the potassium channel: molecular basis of K+ conduction and selectivity.
Science, 280, 1998
|
|
1BYW
 
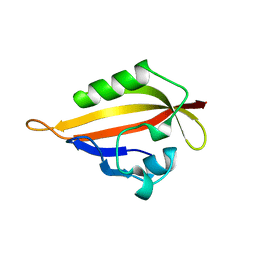 | |
7N0K
 
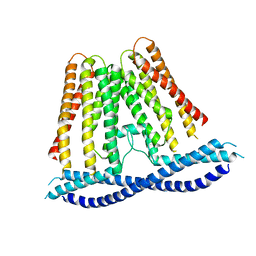 | |
7N0L
 
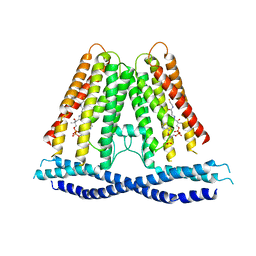 | |
2A79
 
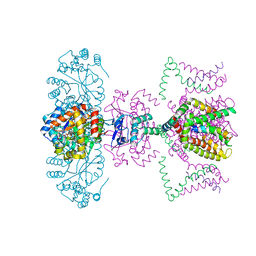 | | Mammalian Shaker Kv1.2 potassium channel- beta subunit complex | | Descriptor: | NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, POTASSIUM ION, Potassium voltage-gated channel subfamily A member 2, ... | | Authors: | Long, S.B, Campbell, E.B, MacKinnon, R. | | Deposit date: | 2005-07-05 | | Release date: | 2005-07-12 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal structure of a mammalian voltage-dependent Shaker family K+ channel.
Science, 309, 2005
|
|
8EMW
 
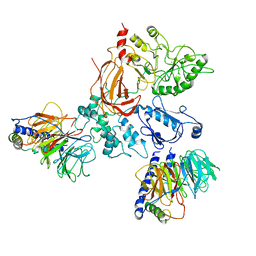 | | Phospholipase C beta 3 (PLCb3) in complex with Gbg on liposomes | | Descriptor: | 1-phosphatidylinositol 4,5-bisphosphate phosphodiesterase beta-3, CALCIUM ION, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Falzone, M.E, MacKinnon, R. | | Deposit date: | 2022-09-28 | | Release date: | 2023-05-24 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | G beta gamma activates PIP2 hydrolysis by recruiting and orienting PLC beta on the membrane surface.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
8EMX
 
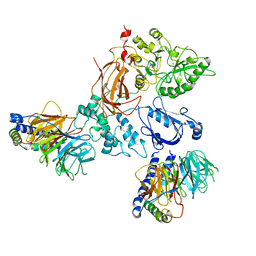 | | Phospholipase C beta 3 (PLCb3) in complex with Gbg on lipid nanodiscs | | Descriptor: | 1-phosphatidylinositol 4,5-bisphosphate phosphodiesterase beta-3, CALCIUM ION, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Falzone, M.E, MacKinnon, R. | | Deposit date: | 2022-09-28 | | Release date: | 2023-05-24 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | G beta gamma activates PIP2 hydrolysis by recruiting and orienting PLC beta on the membrane surface.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
