1D5R
 
 | | Crystal Structure of the PTEN Tumor Suppressor | | Descriptor: | L(+)-TARTARIC ACID, PHOSPHOINOSITIDE PHOSPHATASE PTEN | | Authors: | Lee, J.O, Yang, H, Georgescu, M.-M, Di Cristofano, A, Pavletich, N.P. | | Deposit date: | 1999-10-11 | | Release date: | 1999-11-04 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of the PTEN tumor suppressor: implications for its phosphoinositide phosphatase activity and membrane association.
Cell(Cambridge,Mass.), 99, 1999
|
|
1GUX
 
 | | RB POCKET BOUND TO E7 LXCXE MOTIF | | Descriptor: | ONCOPROTEIN, RETINOBLASTOMA PROTEIN | | Authors: | Lee, J.O, Russo, A.A, Pavletich, N.P. | | Deposit date: | 1997-11-15 | | Release date: | 1998-12-02 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structure of the retinoblastoma tumour-suppressor pocket domain bound to a peptide from HPV E7.
Nature, 391, 1998
|
|
2Z82
 
 | | Crystal structure of the TLR1-TLR2 heterodimer induced by binding of a tri-acylated lipopeptide | | Descriptor: | (2R)-3-{[(2R)-2-AMINO-3-HYDROXYPROPYL]THIO}PROPANE-1,2-DIYL DIHEXADECANOATE, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Lee, J.O, Jin, M.S, Kim, S.E, Heo, J.Y. | | Deposit date: | 2007-08-30 | | Release date: | 2007-10-02 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal Structure of the TLR1-TLR2 Heterodimer Induced by Binding of a Tri-Acylated Lipopeptide
Cell(Cambridge,Mass.), 130, 2007
|
|
2Z80
 
 | | Crystal structure of the TLR1-TLR2 heterodimer induced by binding of a tri-acylated lipopeptide | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Toll-like receptor 2, Variable lymphocyte receptor B | | Authors: | Lee, J.O, Jin, M.S, Kim, S.E, Heo, J.Y. | | Deposit date: | 2007-08-30 | | Release date: | 2007-09-25 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structure of the TLR1-TLR2 Heterodimer Induced by Binding of a Tri-Acylated Lipopeptide
Cell(Cambridge,Mass.), 130, 2007
|
|
2Z7X
 
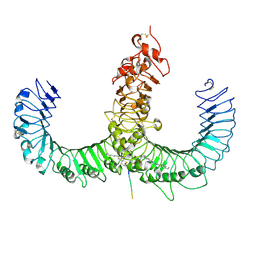 | | Crystal structure of the TLR1-TLR2 heterodimer induced by binding of a tri-acylated lipopeptide | | Descriptor: | (2S)-3-hydroxypropane-1,2-diyl dihexadecanoate, 2-acetamido-2-deoxy-beta-D-glucopyranose, PALMITIC ACID, ... | | Authors: | Lee, J.O, Jin, M.S, Kim, S.E, Heo, J.Y. | | Deposit date: | 2007-08-29 | | Release date: | 2007-10-02 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal Structure of the TLR1-TLR2 Heterodimer Induced by Binding of a Tri-Acylated Lipopeptide
Cell(Cambridge,Mass.), 130, 2007
|
|
2Z81
 
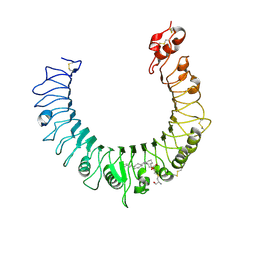 | | Crystal structure of the TLR1-TLR2 heterodimer induced by binding of a tri-acylated lipopeptide | | Descriptor: | (2R)-3-{[(2S)-3-HYDROXY-2-(PALMITOYLAMINO)PROPYL]THIO}PROPANE-1,2-DIYL DIHEXADECANOATE, 2-acetamido-2-deoxy-beta-D-glucopyranose, Toll-like receptor 2, ... | | Authors: | Lee, J.O, Jin, M.S, Kim, S.E, Heo, J.Y. | | Deposit date: | 2007-08-30 | | Release date: | 2007-10-02 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structure of the TLR1-TLR2 Heterodimer Induced by Binding of a Tri-Acylated Lipopeptide
Cell(Cambridge,Mass.), 130, 2007
|
|
6K65
 
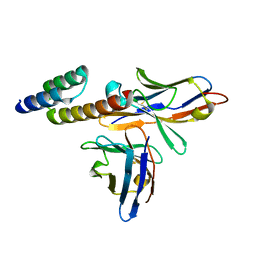 | | Application of anti-helix antibodies in protein structure determination (9014-1P4B) | | Descriptor: | 1P4B variable heavy chain, 1P4B variable light chain, Immunoglobulin G-binding protein A | | Authors: | Lee, J.O, Jin, M.S, Kim, J.W, Kim, S, Lee, H, Cho, G.Y. | | Deposit date: | 2019-06-01 | | Release date: | 2019-08-14 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Application of antihelix antibodies in protein structure determination.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
6K6B
 
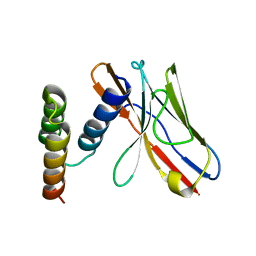 | | Application of anti-helix antibodies in protein structure determination (8496-3LRH) | | Descriptor: | 3LRH intrabody, Protein A | | Authors: | Lee, J.O, Jin, M.S, Kim, J.W, Kim, S, Lee, H, Cho, G.Y. | | Deposit date: | 2019-06-02 | | Release date: | 2019-08-14 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Application of antihelix antibodies in protein structure determination.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
6K3M
 
 | | Application of anti-helix antibodies in protein structure determination (8189-3LRH) | | Descriptor: | 3LRH intrabody, SpA IgG-binding domain protein,Protein A | | Authors: | Lee, J.O, Jin, M.S, Kim, J.W, Kim, S, Lee, H, Cho, G.Y. | | Deposit date: | 2019-05-20 | | Release date: | 2019-08-14 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Application of antihelix antibodies in protein structure determination.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
6K67
 
 | | Application of anti-helix antibodies in protein structure determination (9011-3LRH) | | Descriptor: | 3LRH introbody, CALCIUM ION, Engineered calmodulin | | Authors: | Lee, J.O, Jin, M.S, Kim, J.W, Kim, S, Lee, H, Cho, G.Y. | | Deposit date: | 2019-06-01 | | Release date: | 2019-08-14 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Application of antihelix antibodies in protein structure determination.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
6K68
 
 | | Application of anti-helix antibodies in protein structure determination (8420-3MNZ) | | Descriptor: | 3MNZ Variable heavy chain, 3MNZ Variable light chain, Protein A | | Authors: | Lee, J.O, Jin, M.S, Kim, J.W, Kim, S, Lee, H, Cho, G.Y. | | Deposit date: | 2019-06-01 | | Release date: | 2019-08-14 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Application of antihelix antibodies in protein structure determination.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
6K64
 
 | | Application of anti-helix antibodies in protein structure determination (8188-3LRH) | | Descriptor: | 3LRH intrabody, Protein A | | Authors: | Lee, J.O, Jin, M.S, Kim, J.W, Kim, S, Lee, H, Cho, G.Y. | | Deposit date: | 2019-06-01 | | Release date: | 2019-08-14 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.933 Å) | | Cite: | Application of antihelix antibodies in protein structure determination.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
6K6A
 
 | | Application of anti-helix antibodies in protein structure determination (8188cys-3LRHcys) | | Descriptor: | 3LRH intrabody, Engineered Protein A | | Authors: | Lee, J.O, Jin, M.S, Kim, J.W, Kim, S, Lee, H, Cho, G.Y. | | Deposit date: | 2019-06-02 | | Release date: | 2019-08-14 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Application of antihelix antibodies in protein structure determination.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
6K69
 
 | | Application of anti-helix antibodies in protein structure determination (9213-3LRH) | | Descriptor: | 3LRH intrabody, Engineered T4 lysozyme | | Authors: | Lee, J.O, Jin, M.S, Kim, J.W, Kim, S, Lee, H, Cho, G.Y. | | Deposit date: | 2019-06-01 | | Release date: | 2019-08-14 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.401 Å) | | Cite: | Application of antihelix antibodies in protein structure determination.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
2O6R
 
 | |
2O6S
 
 | |
2O6Q
 
 | |
3Q8W
 
 | | A b-aminoacyl containing thiazolidine derivative and DPPIV complex | | Descriptor: | Dipeptidyl peptidase 4, N-(4-{[({(2R)-3-[(3R)-3-amino-4-(2,4,5-trifluorophenyl)butanoyl]-1,3-thiazolidin-2-yl}carbonyl)amino]methyl}phenyl)-D-valine | | Authors: | Lee, J.O, Song, D.H. | | Deposit date: | 2011-01-07 | | Release date: | 2011-03-16 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (3.64 Å) | | Cite: | Discovery of b-aminoacyl containing thiazolidine derivatives as potent and selective dipeptidyl peptidase IV inhibitors
To be Published
|
|
5X3F
 
 | | Crystal structure of the YgjG-Protein A-Zpa963-PKA catalytic domain | | Descriptor: | Putrescine aminotransferase,Immunoglobulin G-binding protein A, Zpa963,cAMP-dependent protein kinase catalytic subunit alpha | | Authors: | Youn, S.J, Kwon, N.Y, Lee, J.H, Kim, J.H, Lee, H, Lee, J.O. | | Deposit date: | 2017-02-05 | | Release date: | 2017-06-28 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.38 Å) | | Cite: | Construction of novel repeat proteins with rigid and predictable structures using a shared helix method.
Sci Rep, 7, 2017
|
|
4XTT
 
 | | Structural Studies of Potassium Transport Protein KtrA Regulator of Conductance of K+ (RCK) C domain in Complex with Cyclic Diadenosine Monophosphate (c-di-AMP) | | Descriptor: | (2R,3R,3aS,5R,7aR,9R,10R,10aS,12R,14aR)-2,9-bis(6-amino-9H-purin-9-yl)octahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8 ]tetraoxadiphosphacyclododecine-3,5,10,12-tetrol 5,12-dioxide, Putative potassium transport protein | | Authors: | Kim, H, Youn, S.J, Kim, S.O, Ko, J, Lee, J.O, Choi, B.S. | | Deposit date: | 2015-01-24 | | Release date: | 2015-05-20 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.708 Å) | | Cite: | Structural Studies of Potassium Transport Protein KtrA Regulator of Conductance of K+ (RCK) C Domain in Complex with Cyclic Diadenosine Monophosphate (c-di-AMP)
J.Biol.Chem., 290, 2015
|
|
8J26
 
 | | CryoEM structure of SARS CoV-2 RBD and Aptamer complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, AM032-4, AM047-6, ... | | Authors: | Rahman, M.S, Jang, S.K, Lee, J.O. | | Deposit date: | 2023-04-14 | | Release date: | 2023-06-21 | | Last modified: | 2023-07-12 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structure-Guided Development of Bivalent Aptamers Blocking SARS-CoV-2 Infection.
Molecules, 28, 2023
|
|
8J1Q
 
 | | CryoEM structure of SARS CoV-2 RBD and Aptamer complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, AM032-0, AM047-0, ... | | Authors: | Rahman, M.S, Jang, S.K, Lee, J.O. | | Deposit date: | 2023-04-13 | | Release date: | 2023-06-21 | | Last modified: | 2023-07-12 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structure-Guided Development of Bivalent Aptamers Blocking SARS-CoV-2 Infection.
Molecules, 28, 2023
|
|
1BI7
 
 | | MECHANISM OF G1 CYCLIN DEPENDENT KINASE INHIBITION FROM THE STRUCTURE OF THE CDK6-P16INK4A TUMOR SUPPRESSOR COMPLEX | | Descriptor: | CYCLIN-DEPENDENT KINASE 6, MULTIPLE TUMOR SUPPRESSOR | | Authors: | Russo, A.A, Tong, L, Lee, J.O, Jeffrey, P.D, Pavletich, N.P. | | Deposit date: | 1998-06-22 | | Release date: | 1999-01-13 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Structural basis for inhibition of the cyclin-dependent kinase Cdk6 by the tumour suppressor p16INK4a.
Nature, 395, 1998
|
|
1BI8
 
 | | MECHANISM OF G1 CYCLIN DEPENDENT KINASE INHIBITION FROM THE STRUCTURES CDK6-P19INK4D INHIBITOR COMPLEX | | Descriptor: | CYCLIN-DEPENDENT KINASE 6, CYCLIN-DEPENDENT KINASE INHIBITOR | | Authors: | Russo, A.A, Tong, L, Lee, J.O, Jeffrey, P.D, Pavletich, N.P. | | Deposit date: | 1998-06-22 | | Release date: | 1999-01-13 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural basis for inhibition of the cyclin-dependent kinase Cdk6 by the tumour suppressor p16INK4a.
Nature, 395, 1998
|
|
3GWJ
 
 | | Crystal structure of Antheraea pernyi arylphorin | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Arylphorin, FORMIC ACID, ... | | Authors: | Ryu, K.S, Lee, J.O, Kwon, T.H, Kim, S. | | Deposit date: | 2009-04-01 | | Release date: | 2009-05-05 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.43 Å) | | Cite: | The presence of monoglucosylated N196-glycan is important for the structural stability of storage protein, arylphorin
Biochem.J., 421, 2009
|
|
