6WO4
 
 | | Structure of Hepatitis C Virus Envelope Glycoprotein E2 core from genotype 6a bound to broadly neutralizing antibody HC11 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Envelope glycoprotein E2, ... | | Authors: | Tzarum, N, Wilson, I.A, Law, M. | | Deposit date: | 2020-04-24 | | Release date: | 2020-08-19 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.349 Å) | | Cite: | An alternate conformation of HCV E2 neutralizing face as an additional vaccine target.
Sci Adv, 6, 2020
|
|
6WO3
 
 | | Structure of Hepatitis C Virus Envelope Glycoprotein E2 core from genotype 6a bound to broadly neutralizing antibody U1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Envelope glycoprotein E2, ... | | Authors: | Tzarum, N, Wilson, I.A, Law, M. | | Deposit date: | 2020-04-24 | | Release date: | 2020-08-19 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.382 Å) | | Cite: | An alternate conformation of HCV E2 neutralizing face as an additional vaccine target.
Sci Adv, 6, 2020
|
|
6WOQ
 
 | | Structure of Hepatitis C Virus Envelope Glycoprotein E2 core from genotype 1a bound to neutralizing antibody HC1AM and non neutralizing antibody E1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Envelope glycoprotein E2, ... | | Authors: | Tzarum, N, Wilson, I.A, Law, M. | | Deposit date: | 2020-04-25 | | Release date: | 2020-08-19 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.667 Å) | | Cite: | An alternate conformation of HCV E2 neutralizing face as an additional vaccine target.
Sci Adv, 6, 2020
|
|
6WO5
 
 | | Structure of Hepatitis C Virus Envelope Glycoprotein E2 core from genotype 1a bound to neutralizing antibody 212.1.1 and non neutralizing antibody E1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Envelope glycoprotein E2, ... | | Authors: | Tzarum, N, Wilson, I.A, Law, M. | | Deposit date: | 2020-04-24 | | Release date: | 2020-08-19 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.619 Å) | | Cite: | An alternate conformation of HCV E2 neutralizing face as an additional vaccine target.
Sci Adv, 6, 2020
|
|
6WOS
 
 | |
6WOR
 
 | |
4G6A
 
 | |
6BKB
 
 | | Structure of Hepatitis C Virus Envelope Glycoprotein E2 core from genotype 6a bound to broadly neutralizing antibody AR3A | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Fab AR3A heavy chain, ... | | Authors: | Tzarum, N, Wilson, I.A, Law, M. | | Deposit date: | 2017-11-08 | | Release date: | 2018-12-26 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.799 Å) | | Cite: | Genetic and structural insights into broad neutralization of hepatitis C virus by human VH1-69 antibodies.
Sci Adv, 5, 2019
|
|
6BKC
 
 | | Structure of Hepatitis C Virus Envelope Glycoprotein E2 core from genotype 6a bound to broadly neutralizing antibody AR3B | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Fab AR3B heavy chain, Fab AR3B light chain, ... | | Authors: | Tzarum, N, Wilson, I.A, Law, M. | | Deposit date: | 2017-11-08 | | Release date: | 2018-12-26 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Genetic and structural insights into broad neutralization of hepatitis C virus by human VH1-69 antibodies.
Sci Adv, 5, 2019
|
|
6BZV
 
 | | Structure of the Hepatitis C virus envelope glycoprotein E2 antigenic region 412-423 bound to the GL precursor of the broadly neutralizing antibody 19B3 | | Descriptor: | 19B3 GL Heavy Chain, 19B3 GL Light Chain, E2 AS412 peptide | | Authors: | Tzarum, N, Aleman, F, Wilson, I.A, Law, M. | | Deposit date: | 2017-12-26 | | Release date: | 2018-06-20 | | Last modified: | 2022-02-02 | | Method: | X-RAY DIFFRACTION (2.654 Å) | | Cite: | Immunogenetic and structural analysis of a class of HCV broadly neutralizing antibodies and their precursors.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
6BKD
 
 | | Structure of Hepatitis C Virus Envelope Glycoprotein E2 core from genotype 6a bound to broadly neutralizing antibody AR3D | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Fab AR3D heavy chain, Fab AR3D light chain, ... | | Authors: | Tzarum, N, Wilson, I.A, Law, M. | | Deposit date: | 2017-11-08 | | Release date: | 2018-12-26 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.25 Å) | | Cite: | Genetic and structural insights into broad neutralization of hepatitis C virus by human VH1-69 antibodies.
Sci Adv, 5, 2019
|
|
6BZW
 
 | | Structure of the Hepatitis C virus envelope glycoprotein E2 antigenic region 412-423 bound to the GL precursor of the broadly neutralizing antibody AP33 | | Descriptor: | AP33 GL Heavy Chain, AP33 GL Light Chain, E2 AS412 peptide | | Authors: | Tzarum, N, Aleman, F, Wilson, I.A, Law, M. | | Deposit date: | 2017-12-26 | | Release date: | 2018-06-20 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Immunogenetic and structural analysis of a class of HCV broadly neutralizing antibodies and their precursors.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
6BZU
 
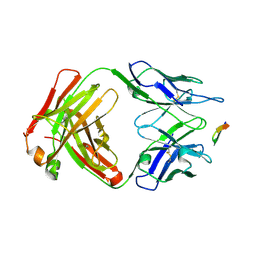 | | Structure of the Hepatitis C virus envelope glycoprotein E2 antigenic region 412-423 bound to the broadly neutralizing antibody 19B3 | | Descriptor: | 19B3 Heavy Chain, 19B3 Light Chain, E2 AS412 peptide | | Authors: | Tzarum, N, Aleman, F, Kong, L, Wilson, I.A, Law, M. | | Deposit date: | 2017-12-26 | | Release date: | 2018-06-20 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Immunogenetic and structural analysis of a class of HCV broadly neutralizing antibodies and their precursors.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
6BZY
 
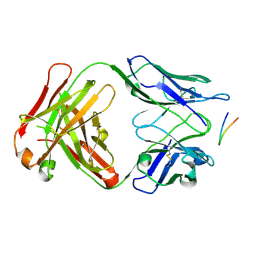 | | Structure of the Hepatitis C virus envelope glycoprotein E2 antigenic region 412-423 bound to the 22D11 broadly neutralizing antibody | | Descriptor: | 22D11 Heavy Chain, 22D11 Light Chain, E2 AS412 peptide | | Authors: | Tzarum, N, Aleman, F, Wilson, I.A, Law, M. | | Deposit date: | 2017-12-26 | | Release date: | 2018-06-20 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Immunogenetic and structural analysis of a class of HCV broadly neutralizing antibodies and their precursors.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
4MWF
 
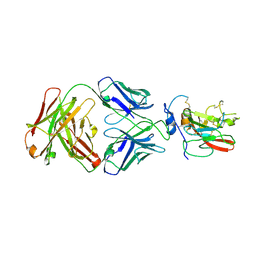 | | Structure of Hepatitis C Virus Envelope Glycoprotein E2 core bound to broadly neutralizing antibody AR3C | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Envelope glycoprotein E2, ... | | Authors: | Kong, L, Wilson, I.A, Law, M. | | Deposit date: | 2013-09-24 | | Release date: | 2013-12-11 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.645 Å) | | Cite: | Hepatitis C virus e2 envelope glycoprotein core structure.
Science, 342, 2013
|
|
4DGV
 
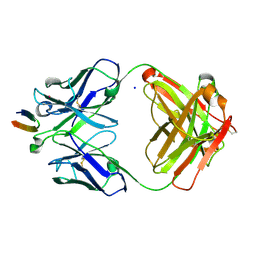 | | Structure of the Hepatitis C virus envelope glycoprotein E2 antigenic region 412-423 bound to the broadly neutralizing antibody HCV1, P2(1) form | | Descriptor: | E2 peptide, HCV1 Heavy Chain, HCV1 Light Chain, ... | | Authors: | Kong, L, Wilson, I.A, Law, M. | | Deposit date: | 2012-01-26 | | Release date: | 2012-05-23 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.805 Å) | | Cite: | Structural basis of hepatitis C virus neutralization by broadly neutralizing antibody HCV1.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
4DGY
 
 | | Structure of the Hepatitis C virus envelope glycoprotein E2 antigenic region 412-423 bound to the broadly neutralizing antibody HCV1, C2 form | | Descriptor: | CHLORIDE ION, E2 peptide, GLYCEROL, ... | | Authors: | Kong, L, Wilson, I.A, Law, M. | | Deposit date: | 2012-01-27 | | Release date: | 2012-05-23 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.798 Å) | | Cite: | Structural basis of hepatitis C virus neutralization by broadly neutralizing antibody HCV1.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
7JTG
 
 | |
7JTF
 
 | |
5VDB
 
 | | Crystal structure of a GNAT superfamily acetyltransferase PA4794 in complex with bisubstrate analog 3 | | Descriptor: | (3R,5S,9R,26S)-1-[(2R,3S,4R,5R)-5-(6-amino-9H-purin-9-yl)-4-hydroxy-3-(phosphonooxy)tetrahydrofuran-2-yl]-3,5,9-trihydroxy-8,8-dimethyl-10,14,20-trioxo-26-({[(phenylacetyl)amino]acetyl}amino)-2,4,6-trioxa-18-thia-11,15,21-triaza-3,5-diphosphaheptacosan-27-oic acid 3,5-dioxide (non-preferred name), SULFATE ION, acetyltransferase PA4794 | | Authors: | Majorek, K.A, Joachimiak, A, Minor, W, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2017-04-01 | | Release date: | 2017-07-26 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Generating enzyme and radical-mediated bisubstrates as tools for investigating Gcn5-related N-acetyltransferases.
FEBS Lett., 591, 2017
|
|
8FSJ
 
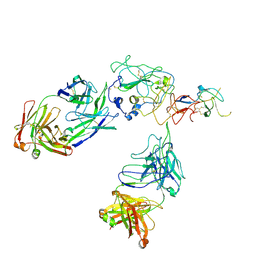 | | Cryo-EM structure of engineered hepatitis C virus E1E2 ectodomain in complex with antibodies AR4A, HEPC74, and IGH520 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, AR4A heavy chain, ... | | Authors: | Metcalf, M.C, Ofek, G. | | Deposit date: | 2023-01-10 | | Release date: | 2023-07-19 | | Method: | ELECTRON MICROSCOPY (3.65 Å) | | Cite: | Structure of engineered hepatitis C virus E1E2 ectodomain in complex with neutralizing antibodies.
Nat Commun, 14, 2023
|
|
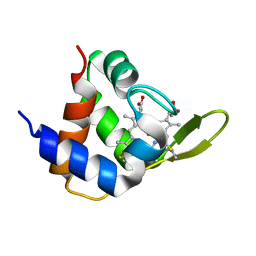
4XXL
 
 | |
5VD6
 
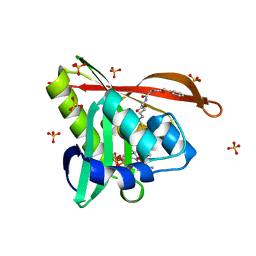 | | Crystal structure of a GNAT superfamily acetyltransferase PA4794 in complex with bisubstrate analog 6 | | Descriptor: | (3R,5S,9R,23S)-1-[(2R,3S,4R,5R)-5-(6-amino-9H-purin-9-yl)-4-hydroxy-3-(phosphonooxy)tetrahydrofuran-2-yl]-3,5,9-trihydroxy-8,8-dimethyl-10,14-dioxo-23-({[(phenylacetyl)amino]acetyl}amino)-2,4,6-trioxa-18-thia-11,15-diaza-3,5-diphosphatetracosan-24-oic acid 3,5-dioxide (non-preferred name), SULFATE ION, acetyltransferase PA4794 | | Authors: | Majorek, K.A, Joachimiak, A, Minor, W, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2017-04-01 | | Release date: | 2017-07-26 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Generating enzyme and radical-mediated bisubstrates as tools for investigating Gcn5-related N-acetyltransferases.
FEBS Lett., 591, 2017
|
|
4EVY
 
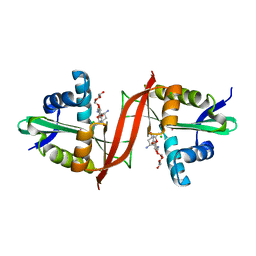 | | Crystal structure of aminoglycoside antibiotic 6'-N-acetyltransferase AAC(6')-Ig from Acinetobacter haemolyticus in complex with tobramycin | | Descriptor: | Aminoglycoside N(6')-acetyltransferase type 1, CHLORIDE ION, POTASSIUM ION, ... | | Authors: | Stogios, P.J, Evdokimova, E, Minasov, G, Yim, V, Courvalin, P, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2012-04-26 | | Release date: | 2012-05-09 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.768 Å) | | Cite: | Structural and Biochemical Characterization of Acinetobacter spp. Aminoglycoside Acetyltransferases Highlights Functional and Evolutionary Variation among Antibiotic Resistance Enzymes.
ACS Infect Dis., 3, 2017
|
|
4F0Y
 
 | | Crystal structure of aminoglycoside antibiotic 6'-N-acetyltransferase AAC(6')-IG from Acinetobacter haemolyticus, apo | | Descriptor: | Aminoglycoside N(6')-acetyltransferase type 1, CHLORIDE ION, GLYCEROL, ... | | Authors: | Stogios, P.J, Evdokimova, E, Dong, A, Minasov, G, Yim, V, Courvalin, P, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2012-05-05 | | Release date: | 2012-05-16 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.56 Å) | | Cite: | Structural and Biochemical Characterization of Acinetobacter spp. Aminoglycoside Acetyltransferases Highlights Functional and Evolutionary Variation among Antibiotic Resistance Enzymes.
ACS Infect Dis., 3, 2017
|
|
