4SGA
 
 | |
5SGA
 
 | |
1APW
 
 | |
1APV
 
 | |
3SGA
 
 | |
3APP
 
 | |
2SGA
 
 | |
2PSG
 
 | |
1APT
 
 | |
1APU
 
 | |
1HAV
 
 | | HEPATITIS A VIRUS 3C PROTEINASE | | Descriptor: | CHLORIDE ION, HEPATITIS A VIRUS 3C PROTEINASE | | Authors: | Bergmann, E.M, James, M.N.G. | | Deposit date: | 1996-10-23 | | Release date: | 1996-12-23 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The refined crystal structure of the 3C gene product from hepatitis A virus: specific proteinase activity and RNA recognition.
J.Virol., 71, 1997
|
|
1QA7
 
 | | CRYSTAL COMPLEX OF THE 3C PROTEINASE FROM HEPATITIS A VIRUS WITH ITS INHIBITOR AND IMPLICATIONS FOR THE POLYPROTEIN PROCESSING IN HAV | | Descriptor: | DIMETHYL SULFOXIDE, GLYCEROL, HAV 3C PROTEINASE, ... | | Authors: | Bergmann, E.M, Cherney, M.M, Mckendrick, J, Vederas, J.C, James, M.N.G. | | Deposit date: | 1999-04-15 | | Release date: | 1999-04-20 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of an inhibitor complex of the 3C proteinase from hepatitis A virus (HAV) and implications for the polyprotein processing in HAV.
Virology, 265, 1999
|
|
1QRP
 
 | | Human pepsin 3A in complex with a phosphonate inhibitor IVA-VAL-VAL-LEU(P)-(O)PHE-ALA-ALA-OME | | Descriptor: | PEPSIN 3A, methyl N-[(2S)-2-({(S)-hydroxy[(1R)-3-methyl-1-{[N-(3-methylbutanoyl)-L-valyl-L-valyl]amino}butyl]phosphoryl}oxy)-3-phenylpropanoyl]-L-alanyl-L-alaninate | | Authors: | Fujinaga, M, Cherney, M.M, Tarasova, N.I, Bartlett, P.A, Hanson, J.E, James, M.N.G. | | Deposit date: | 1999-06-15 | | Release date: | 1999-06-18 | | Last modified: | 2012-12-12 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Structural study of the complex between human pepsin and a phosphorus-containing peptidic -transition-state analog.
Acta Crystallogr.,Sect.D, 56, 2000
|
|
2A5K
 
 | | Crystal structures of SARS coronavirus main peptidase inhibited by an aza-peptide epoxide in space group P212121 | | Descriptor: | (5S,8S,14R)-ETHYL 11-(3-AMINO-3-OXOPROPYL)-8-BENZYL-14-HYDROXY-5-ISOBUTYL-3,6,9,12-TETRAOXO-1-PHENYL-2-OXA-4,7,10,11-TETRAAZAPENTADECAN-15-OATE, 3C-like peptidase | | Authors: | Lee, T.-W, Cherney, M.M, Huitema, C, Liu, J, James, K.E, Powers, J.C, Eltis, L.D, James, M.N. | | Deposit date: | 2005-06-30 | | Release date: | 2005-10-25 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structures of the Main Peptidase from the SARS Coronavirus Inhibited by a Substrate-like Aza-peptide Epoxide
J.Mol.Biol., 353, 2005
|
|
5TNC
 
 | |
2A5A
 
 | | Crystal structure of unbound SARS coronavirus main peptidase in the space group C2 | | Descriptor: | 1,2-ETHANEDIOL, 3C-like peptidase, CHLORIDE ION | | Authors: | Lee, T.-W, Cherney, M.M, Huitema, C, Liu, J, James, K.E, Powers, J.C, Eltis, L.D, James, M.N. | | Deposit date: | 2005-06-30 | | Release date: | 2005-10-25 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | Crystal Structures of the Main Peptidase from the SARS Coronavirus Inhibited by a Substrate-like Aza-peptide Epoxide
J.Mol.Biol., 353, 2005
|
|
3SGB
 
 | | STRUCTURE OF THE COMPLEX OF STREPTOMYCES GRISEUS PROTEASE B AND THE THIRD DOMAIN OF THE TURKEY OVOMUCOID INHIBITOR AT 1.8 ANGSTROMS RESOLUTION | | Descriptor: | PROTEINASE B (SGPB), TURKEY OVOMUCOID INHIBITOR (OMTKY3) | | Authors: | Read, R.J, Fujinaga, M, Sielecki, A.R, James, M.N.G. | | Deposit date: | 1983-01-21 | | Release date: | 1983-07-12 | | Last modified: | 2017-11-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structure of the complex of Streptomyces griseus protease B and the third domain of the turkey ovomucoid inhibitor at 1.8-A resolution.
Biochemistry, 22, 1983
|
|
1QS8
 
 | | Crystal structure of the P. vivax aspartic proteinase plasmepsin complexed with the inhibitor pepstatin A | | Descriptor: | ACETATE ION, PEPSTATIN A, PLASMEPSIN | | Authors: | Khazanovich Bernstein, N, Cherney, M.M, Yowell, C.A, Dame, J.B, James, M.N.G. | | Deposit date: | 1999-06-25 | | Release date: | 1999-07-07 | | Last modified: | 2013-02-27 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural insights into the activation of P. vivax plasmepsin.
J.Mol.Biol., 329, 2003
|
|
2A5I
 
 | | Crystal structures of SARS coronavirus main peptidase inhibited by an aza-peptide epoxide in the space group C2 | | Descriptor: | (5S,8S,14R)-ETHYL 11-(3-AMINO-3-OXOPROPYL)-8-BENZYL-14-HYDROXY-5-ISOBUTYL-3,6,9,12-TETRAOXO-1-PHENYL-2-OXA-4,7,10,11-TETRAAZAPENTADECAN-15-OATE, 1,2-ETHANEDIOL, 3C-like peptidase, ... | | Authors: | Lee, T.-W, Cherney, M.M, Huitema, C, Liu, J, James, K.E, Powers, J.C, Eltis, L.D, James, M.N. | | Deposit date: | 2005-06-30 | | Release date: | 2005-10-25 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Crystal Structures of the Main Peptidase from the SARS Coronavirus Inhibited by a Substrate-like Aza-peptide Epoxide
J.Mol.Biol., 353, 2005
|
|
2AQ6
 
 | | X-ray crystal structure of mycobacterium tuberculosis pyridoxine 5'-phosphate oxidase complexed with pyridoxal 5'-phosphate at 1.7 a resolution | | Descriptor: | PYRIDOXAL-5'-PHOSPHATE, PYRIDOXINE 5'-PHOSPHATE OXIDASE | | Authors: | Biswal, B.K, Cherney, M.M, Wang, M, Garen, C, James, M.N, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2005-08-17 | | Release date: | 2005-08-30 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structures of Mycobacterium tuberculosispyridoxine 5'-phosphate oxidase and its complexes with flavin mononucleotide and pyridoxal 5'-phosphate.
Acta Crystallogr.,Sect.D, 61, 2005
|
|
2CI2
 
 | |
4DGI
 
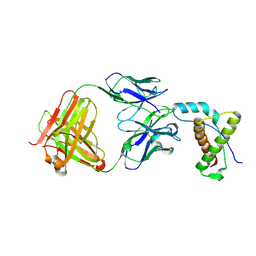 | | Structure of POM1 FAB fragment complexed with human PrPc Fragment 120-230 | | Descriptor: | Major prion protein, POM1 Fab Heavy chain, POM1 Fab Light chain, ... | | Authors: | Baral, P.K, Wieland, B, Swayampakula, M, James, M.N. | | Deposit date: | 2012-01-26 | | Release date: | 2012-10-31 | | Last modified: | 2013-01-23 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural studies on the folded domain of the human prion protein bound to the Fab fragment of the antibody POM1.
Acta Crystallogr.,Sect.D, 68, 2012
|
|
2ATO
 
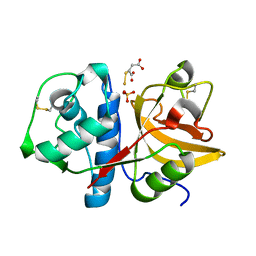 | | Crystal structure of Human Cathepsin K in complex with myocrisin | | Descriptor: | (S)-(1,2-DICARBOXYETHYLTHIO)GOLD, Cathepsin K, SULFATE ION | | Authors: | Weidauer, E, Yasuda, Y, Biswal, B.K, Kerr, L.D, Cherney, M.M, Gordon, R.E, James, M.N.G, Bromme, D. | | Deposit date: | 2005-08-25 | | Release date: | 2006-08-29 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Effects of disease-modifying anti-rheumatic drugs (DMARDs) on the activities of rheumatoid arthritis-associated cathepsins K and S.
Biol.Chem., 388, 2007
|
|
2ALP
 
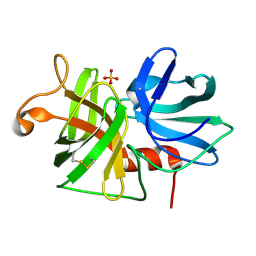 | | REFINED STRUCTURE OF ALPHA-LYTIC PROTEASE AT 1.7 ANGSTROMS RESOLUTION. ANALYSIS OF HYDROGEN BONDING AND SOLVENT STRUCTURE | | Descriptor: | ALPHA-LYTIC PROTEASE, SULFATE ION | | Authors: | Fujinaga, M, Delbaere, L.T.J, Brayer, G.D, James, M.N.G. | | Deposit date: | 1985-03-07 | | Release date: | 1985-07-17 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Refined structure of alpha-lytic protease at 1.7 A resolution. Analysis of hydrogen bonding and solvent structure.
J.Mol.Biol., 184, 1985
|
|
2HAL
 
 | | An episulfide cation (thiiranium ring) trapped in the active site of HAV 3C proteinase inactivated by peptide-based ketone inhibitors | | Descriptor: | Hepatitis A Protease 3C, N-ACETYL-LEUCYL-PHENYLALANYL-PHENYLALANYL-GLUTAMATE-FLUOROMETHYLKETONE INHIBITOR, N-[(BENZYLOXY)CARBONYL]-L-ALANINE | | Authors: | Yin, J, Cherney, M.M, Bergmann, E.M, James, M.N. | | Deposit date: | 2006-06-13 | | Release date: | 2006-08-08 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | An Episulfide Cation (Thiiranium Ring) Trapped in the Active Site of HAV 3C Proteinase Inactivated by Peptide-based Ketone Inhibitors.
J.Mol.Biol., 361, 2006
|
|
