4IDU
 
 | |
4IDX
 
 | |
4N4R
 
 | | Structure basis of lipopolysaccharide biogenesis | | Descriptor: | CACODYLATE ION, LPS-assembly lipoprotein LptE, LPS-assembly protein LptD, ... | | Authors: | Dong, H, Xiang, Q, Wang, Z, Paterson, N.G, He, C, Zhang, Y, Wang, W, Dong, C. | | Deposit date: | 2013-10-08 | | Release date: | 2014-06-25 | | Last modified: | 2014-07-16 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural basis for outer membrane lipopolysaccharide insertion.
Nature, 511, 2014
|
|
3E3R
 
 | |
5HDE
 
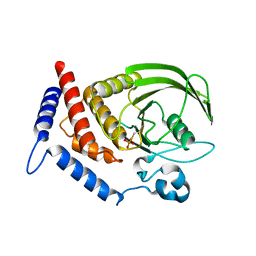 | | Crystal Structure of PTPN12 Catalytic Domain | | Descriptor: | PHOSPHATE ION, Tyrosine-protein phosphatase non-receptor type 12 | | Authors: | Dong, H, Li, S, Shi, J. | | Deposit date: | 2016-01-05 | | Release date: | 2017-01-18 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Crystal Structure of PTPN12 Catalytic Domain
To Be Published
|
|
7N09
 
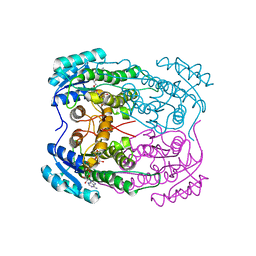 | |
7DST
 
 | | FMDV capsid in complex with M170 Nab | | Descriptor: | M170 Nab, VP1 of O type FMDV capsid, VP2 of O type FMDV capsid, ... | | Authors: | Dong, H, Liu, P. | | Deposit date: | 2021-01-02 | | Release date: | 2021-03-10 | | Last modified: | 2022-05-25 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural and molecular basis for foot-and-mouth disease virus neutralization by two potent protective antibodies.
Protein Cell, 13, 2022
|
|
7DSS
 
 | | Complex of FMDV and M8 Nab | | Descriptor: | M8 Nab, VP1 of O type FMDV capsid, VP2 of O-type FMDV capsid, ... | | Authors: | Dong, H, Liu, P. | | Deposit date: | 2021-01-02 | | Release date: | 2021-03-31 | | Last modified: | 2022-05-25 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Structural and molecular basis for foot-and-mouth disease virus neutralization by two potent protective antibodies.
Protein Cell, 13, 2022
|
|
7ENO
 
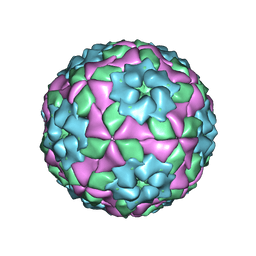 | | Mutant strain M3 of foot-and-mouth disease virus type O | | Descriptor: | VP1 of O type FMDV capsid, VP2 of O type FMDV capsid, VP3 of O type FMDV capsid, ... | | Authors: | Dong, H, Lu, Y. | | Deposit date: | 2021-04-18 | | Release date: | 2021-06-02 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.15 Å) | | Cite: | A Heat-Induced Mutation on VP1 of Foot-and-Mouth Disease Virus Serotype O Enhanced Capsid Stability and Immunogenicity.
J.Virol., 95, 2021
|
|
7ENP
 
 | | wild type of O type Foot-and-mouth disease virus | | Descriptor: | VP1 of O type FMDV capsid protein, VP2 of O type FMDV capsid protein, VP3 of O type FMDV capsid protein, ... | | Authors: | Dong, H, Lu, Y. | | Deposit date: | 2021-04-18 | | Release date: | 2021-06-02 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | A Heat-Induced Mutation on VP1 of Foot-and-Mouth Disease Virus Serotype O Enhanced Capsid Stability and Immunogenicity.
J.Virol., 95, 2021
|
|
6A2F
 
 | |
5YYC
 
 | | Crystal structure of alanine racemase from Bacillus pseudofirmus (OF4) | | Descriptor: | Alanine racemase, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Dong, H, Hu, T.T, He, G.Z, Lu, D.R, Qi, J.X, Dou, Y.S, Long, W, He, X, Su, D, Ju, J.S. | | Deposit date: | 2017-12-08 | | Release date: | 2019-01-02 | | Method: | X-RAY DIFFRACTION (1.801 Å) | | Cite: | Structural features and kinetic characterization of alanine racemase from Bacillus pseudofirmus OF4.
Biochem. Biophys. Res. Commun., 497, 2018
|
|
5L75
 
 | | A protein structure | | Descriptor: | FIG000906: Predicted Permease, FIG000988: Predicted permease, Lipopolysaccharide ABC transporter, ... | | Authors: | Dong, C, Dong, H, Zhang, Z, Paterson, N, Tang, X. | | Deposit date: | 2016-06-01 | | Release date: | 2017-08-16 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3.7 Å) | | Cite: | Structural and functional insights into the lipopolysaccharide ABC transporter LptB2FG.
Nat Commun, 8, 2017
|
|
5I5H
 
 | | Ecoli global domain 245-586 | | Descriptor: | Inner membrane protein YejM | | Authors: | Dong, C, Dong, H. | | Deposit date: | 2016-02-15 | | Release date: | 2016-08-17 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structural insights into cardiolipin transfer from the Inner membrane to the outer membrane by PbgA in Gram-negative bacteria.
Sci Rep, 6, 2016
|
|
5I5D
 
 | | Salmonella global domain 245 | | Descriptor: | Inner membrane protein YejM | | Authors: | Dong, C, Dong, H. | | Deposit date: | 2016-02-15 | | Release date: | 2017-04-19 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Structural insights into cardiolipin transfer from the Inner membrane to the outer membrane by PbgA in Gram-negative bacteria.
Sci Rep, 6, 2016
|
|
5I5F
 
 | | Salmonella global domain 191 | | Descriptor: | Inner membrane protein YejM | | Authors: | Dong, C, Dong, H. | | Deposit date: | 2016-02-15 | | Release date: | 2016-08-17 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Structural insights into cardiolipin transfer from the Inner membrane to the outer membrane by PbgA in Gram-negative bacteria.
Sci Rep, 6, 2016
|
|
2L0J
 
 | | Solid State NMR structure of the M2 proton channel from Influenza A Virus in hydrated lipid bilayer | | Descriptor: | Matrix protein 2 | | Authors: | Sharma, M, Yi, M, Dong, H, Qin, H, Peterson, E, Busath, D.D, Zhou, H.X, Cross, T.A. | | Deposit date: | 2010-07-08 | | Release date: | 2010-11-03 | | Last modified: | 2024-05-01 | | Method: | SOLID-STATE NMR | | Cite: | Insight into the mechanism of the influenza a proton channel from a structure in a lipid bilayer.
Science, 330, 2010
|
|
8BO2
 
 | | BAM-EspP complex structure with BamA-S425C/EspP-S1299C mutations in nanodisc | | Descriptor: | Outer membrane protein assembly factor BamA, Outer membrane protein assembly factor BamB, Outer membrane protein assembly factor BamC, ... | | Authors: | Shen, C, Chang, S, Luo, Q, Zhang, Z, Xie, T, Luo, B, Lu, G, Zhu, X, Wei, X, Dong, C, Zhou, R, Zhang, X, Tang, X, Dong, H. | | Deposit date: | 2022-11-14 | | Release date: | 2023-04-26 | | Last modified: | 2023-05-24 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural basis of BAM-mediated outer membrane beta-barrel protein assembly.
Nature, 617, 2023
|
|
8BNZ
 
 | | BAM-EspP complex structure with BamA-G431C/EspP-N1293C mutations in nanodisc | | Descriptor: | Outer membrane protein assembly factor BamA, Outer membrane protein assembly factor BamB, Outer membrane protein assembly factor BamC, ... | | Authors: | Shen, C, Chang, S, Luo, Q, Zhang, Z, Xie, T, Luo, B, Lu, G, Zhu, X, Wei, X, Dong, C, Zhou, R, Zhang, X, Tang, X, Dong, H. | | Deposit date: | 2022-11-14 | | Release date: | 2023-04-26 | | Last modified: | 2023-05-24 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structural basis of BAM-mediated outer membrane beta-barrel protein assembly.
Nature, 617, 2023
|
|
4QC2
 
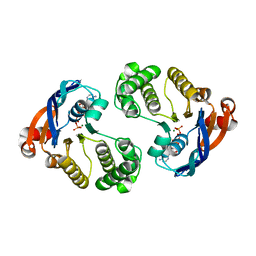 | | Crystal structure of lipopolysaccharide transport protein LptB in complex with ATP and Magnesium ions | | Descriptor: | ABC transporter related protein, ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION | | Authors: | Wang, Z, Xiang, Q, Zhu, X, Dong, H, He, C, Wang, H, Zhang, Y, Wang, W, Dong, C. | | Deposit date: | 2014-05-09 | | Release date: | 2014-10-22 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | Structural and functional studies of conserved nucleotide-binding protein LptB in lipopolysaccharide transport.
Biochem.Biophys.Res.Commun., 452, 2014
|
|
4G9Z
 
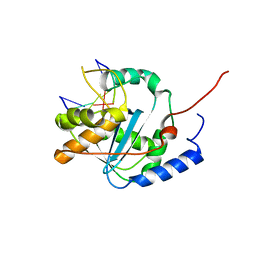 | | Lassa nucleoprotein with dsRNA reveals novel mechanism for immune suppression | | Descriptor: | Nucleoprotein, RNA (5'-R(P*GP*CP*CP*C)-3'), RNA (5'-R(P*GP*GP*GP*C)-3'), ... | | Authors: | Jiang, X, Huang, Q, Wang, W, Dong, H, Ly, H, Liang, Y, Dong, C, Scottish Structural Proteomics Facility (SSPF) | | Deposit date: | 2012-07-24 | | Release date: | 2013-04-03 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Structures of Arenaviral Nucleoproteins with Triphosphate dsRNA Reveal a Unique Mechanism of Immune Suppression.
J.Biol.Chem., 288, 2013
|
|
4GV6
 
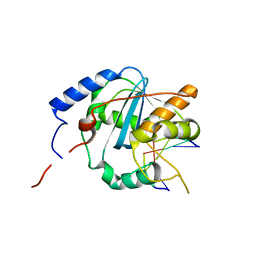 | | Structures of Lassa and Tacaribe viral nucleoproteins with or without 5 triphosphate dsRNA substrate reveal a unique 3 -5 exoribonuclease mechanism to suppress type I interferon production | | Descriptor: | MANGANESE (II) ION, Nucleoprotein, RNA (5'-R(*(GTP)P*GP*GP*C)-3'), ... | | Authors: | Jiang, X, Huang, Q, Wang, W, Dong, H, Ly, H, Liang, Y, Dong, C. | | Deposit date: | 2012-08-30 | | Release date: | 2013-05-01 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Structures of Arenaviral Nucleoproteins with Triphosphate dsRNA Reveal a Unique Mechanism of Immune Suppression.
J.Biol.Chem., 288, 2013
|
|
4GVE
 
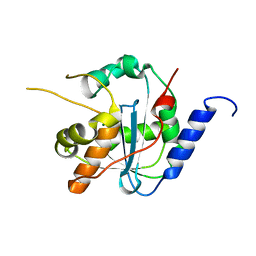 | | Tacaribe nucleoprotein structure | | Descriptor: | MAGNESIUM ION, Nucleoprotein, ZINC ION | | Authors: | Jiang, X, Huang, Q, Wang, W, Dong, H, Ly, H, Liang, Y, Dong, C. | | Deposit date: | 2012-08-30 | | Release date: | 2013-05-01 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Structures of Arenaviral Nucleoproteins with Triphosphate dsRNA Reveal a Unique Mechanism of Immune Suppression.
J.Biol.Chem., 288, 2013
|
|
4GV9
 
 | | Lassa nucleoprotein C-terminal domain in complex with triphosphated dsRNA soaking for 5 min | | Descriptor: | MANGANESE (II) ION, Nucleoprotein, RNA (5'-R(*(GTP)P*GP*GP*C)-3'), ... | | Authors: | Jiang, X, Huang, Q, Wang, W, Dong, H, Ly, H, Liang, Y, Dong, C. | | Deposit date: | 2012-08-30 | | Release date: | 2013-05-01 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.46 Å) | | Cite: | Structures of Arenaviral Nucleoproteins with Triphosphate dsRNA Reveal a Unique Mechanism of Immune Suppression.
J.Biol.Chem., 288, 2013
|
|
4MIW
 
 | | High-resolution structure of the N-terminal endonuclease domain of the Lassa virus L polymerase | | Descriptor: | MAGNESIUM ION, RNA-directed RNA polymerase L | | Authors: | Wallat, G.D, Huang, Q, Wang, W, Dong, H, Ly, H, Liang, Y, Dong, C. | | Deposit date: | 2013-09-02 | | Release date: | 2014-03-19 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | High-resolution structure of the N-terminal endonuclease domain of the lassa virus L polymerase in complex with magnesium ions.
Plos One, 9, 2014
|
|
