6GY5
 
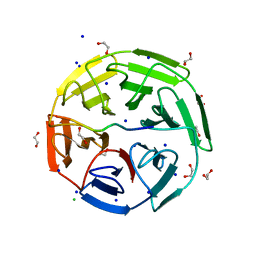 | | Crystal structure of the kelch domain of human KLHL20 in complex with DAPK1 peptide | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Death-associated protein kinase 1, ... | | Authors: | Chen, Z, Hozjan, V, Strain-Damerell, C, Williams, E, Wang, D, Cooper, C.D.O, Sanvitale, C.E, Fairhead, M, Carpenter, E.P, Pike, A.C.W, Krojer, T, Srikannathasan, V, Sorrell, F, Johansson, C, Mathea, S, Burgess-Brown, N, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Bullock, A.N. | | Deposit date: | 2018-06-28 | | Release date: | 2018-08-08 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.086 Å) | | Cite: | Structural Basis for Recruitment of DAPK1 to the KLHL20 E3 Ligase.
Structure, 27, 2019
|
|
6HRL
 
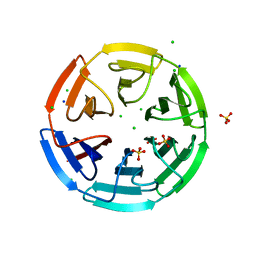 | | Crystal structure of the Kelch domain of human KLHL17 | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Kelch-like protein 17, ... | | Authors: | Chen, Z, Williams, E, Sorrell, F.J, Newman, J.A, Shrestha, L, Burgess-Brown, N, von Delft, F, Arrowsmith, F, Edwards, C.H, Bountra, C, Bullock, A.N. | | Deposit date: | 2018-09-27 | | Release date: | 2018-10-17 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of the Kelch domain of human KLHL17
To Be Published
|
|
5NKP
 
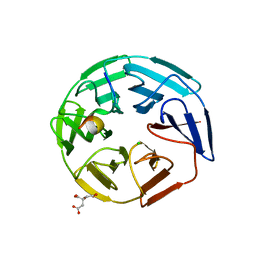 | | Crystal structure of the human KLHL3 Kelch domain in complex with a WNK3 peptide | | Descriptor: | CHLORIDE ION, CITRIC ACID, Kelch-like protein 3, ... | | Authors: | Chen, Z, Sorrell, F.J, Pinkas, D.M, Williams, E, Mathea, S, Goubin, S, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Burgess-Brown, N, Bountra, C, Bullock, A. | | Deposit date: | 2017-03-31 | | Release date: | 2017-05-03 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of the human KLHL3 Kelch domain in complex with a WNK3 peptide
To Be Published
|
|
1HSH
 
 | |
1HSI
 
 | |
1HSG
 
 | |
4XGA
 
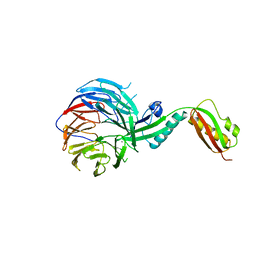 | | Crystal structure of BamB and BamA P3-5 complex from E.coli | | Descriptor: | CALCIUM ION, Outer membrane protein assembly factor BamA, Outer membrane protein assembly factor BamB | | Authors: | Chen, Z, Zhan, L.H, Dong, C, Gao, Z.Q, Dong, Y.H. | | Deposit date: | 2014-12-30 | | Release date: | 2016-01-20 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structural basis for the interaction of BamB with the POTRA3-4 domains of BamA.
Acta Crystallogr D Struct Biol, 72, 2016
|
|
1AHT
 
 | |
1C6V
 
 | | SIV INTEGRASE (CATALYTIC DOMAIN + DNA BIDING DOMAIN COMPRISING RESIDUES 50-293) MUTANT WITH PHE 185 REPLACED BY HIS (F185H) | | Descriptor: | PROTEIN (SIU89134), PROTEIN (SIV INTEGRASE) | | Authors: | Chen, Z, Yan, Y, Munshi, S, Li, Y, Zruygay-Murphy, J, Xu, B, Witmer, M, Felock, P, Wolfe, A, Sardana, V, Emini, E.A, Hazuda, D, Kuo, L.C. | | Deposit date: | 1999-12-21 | | Release date: | 2000-12-27 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | X-ray structure of simian immunodeficiency virus integrase containing the core and C-terminal domain (residues 50-293)--an initial glance of the viral DNA binding platform.
J.Mol.Biol., 296, 2000
|
|
8D6T
 
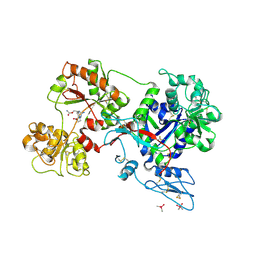 | | Rana catesbeiana saxiphilin mutant - Y558I:STX (co-crystal) | | Descriptor: | CACODYLATE ION, Saxiphilin, [(3aS,4R,10aS)-2,6-diamino-10,10-dihydroxy-3a,4,9,10-tetrahydro-3H,8H-pyrrolo[1,2-c]purin-4-yl]methyl carbamate | | Authors: | Chen, Z, Zakrzewska, S, Minor, D.L. | | Deposit date: | 2022-06-06 | | Release date: | 2022-11-02 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Definition of a saxitoxin (STX) binding code enables discovery and characterization of the anuran saxiphilin family.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
8D6P
 
 | |
8D6Q
 
 | |
8D6S
 
 | | Rana catesbeiana saxiphilin mutant - Y558A:STX (co-crystal) | | Descriptor: | Saxiphilin, [(3aS,4R,10aS)-2,6-diamino-10,10-dihydroxy-3a,4,9,10-tetrahydro-3H,8H-pyrrolo[1,2-c]purin-4-yl]methyl carbamate | | Authors: | Chen, Z, Zakrzewska, S, Minor, D.L. | | Deposit date: | 2022-06-06 | | Release date: | 2022-11-02 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Definition of a saxitoxin (STX) binding code enables discovery and characterization of the anuran saxiphilin family.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
8D6U
 
 | | Rana catesbeiana saxiphilin:F-STX (soaked) | | Descriptor: | (2P)-4-({6-[({[(3aS,4R,7R,10aS)-2,6-diamino-10,10-dihydroxy-3a,4,9,10-tetrahydro-3H,8H-pyrrolo[1,2-c]purin-4-yl]methoxy}carbonyl)amino]hexyl}carbamoyl)-2-{[4aP,9(9a)P]-6-hydroxy-3-oxo-3H-xanthen-9-yl}benzoic acid, Saxiphilin | | Authors: | Chen, Z, Zakrzewska, S, Minor, D.L. | | Deposit date: | 2022-06-06 | | Release date: | 2022-11-02 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Definition of a saxitoxin (STX) binding code enables discovery and characterization of the anuran saxiphilin family.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
3KZ1
 
 | | Crystal Structure of the Complex of PDZ-RhoGEF DH/PH domains with GTP-gamma-S Activated RhoA | | Descriptor: | 5'-GUANOSINE-DIPHOSPHATE-MONOTHIOPHOSPHATE, MAGNESIUM ION, Rho guanine nucleotide exchange factor 11, ... | | Authors: | Chen, Z, Sternweis, P.C, Sprang, S.R. | | Deposit date: | 2009-12-07 | | Release date: | 2010-04-28 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Activated RhoA binds to the pleckstrin homology (PH) domain of PDZ-RhoGEF, a potential site for autoregulation.
J.Biol.Chem., 285, 2010
|
|
1EKM
 
 | | CRYSTAL STRUCTURE AT 2.5 A RESOLUTION OF ZINC-SUBSTITUTED COPPER AMINE OXIDASE OF HANSENULA POLYMORPHA EXPRESSED IN ESCHERICHIA COLI | | Descriptor: | COPPER AMINE OXIDASE, ZINC ION | | Authors: | Chen, Z, Schwartz, B, Williams, N.K, Li, R, Klinman, J.P, Mathews, F.S. | | Deposit date: | 2000-03-09 | | Release date: | 2000-08-23 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure at 2.5 A resolution of zinc-substituted copper amine oxidase of Hansenula polymorpha expressed in Escherichia coli.
Biochemistry, 39, 2000
|
|
1FVY
 
 | |
1SHZ
 
 | | Crystal Structure of the p115RhoGEF rgRGS Domain in A Complex with Galpha(13):Galpha(i1) Chimera | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, Guanine Nucleotide-Binding Protein Galpha(13):Galpha(i1) Chimera, MAGNESIUM ION, ... | | Authors: | Chen, Z, Singer, W.D, Sternweis, P.C, Sprang, S.R. | | Deposit date: | 2004-02-26 | | Release date: | 2005-01-18 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Structure of the p115RhoGEF rgRGS domain-Galpha13/i1 chimera complex suggests convergent evolution of a GTPase activator.
Nat.Struct.Mol.Biol., 12, 2005
|
|
6TTK
 
 | | Crystal structure of the kelch domain of human KLHL12 in complex with DVL1 peptide | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, DVL1, ... | | Authors: | Chen, Z, Williams, E, Pike, A.C.W, Strain-Damerell, C, Wang, D, Chalk, R, Burgess-Brown, N, Krojer, T, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Bullock, A.N. | | Deposit date: | 2019-12-27 | | Release date: | 2020-02-12 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.383 Å) | | Cite: | Identification of a PGXPP degron motif in dishevelled and structural basis for its binding to the E3 ligase KLHL12.
Open Biology, 10, 2020
|
|
8JT8
 
 | | Crystal structure of 5-HT2AR in complex with (R)-IHCH-7179 | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 1-(4-fluorophenyl)-4-[(7R)-2,5,11-triazatetracyclo[7.6.1.0^2,7.0^12,16]hexadeca-1(15),9,12(16),13-tetraen-5-yl]butan-1-one, 5-hydroxytryptamine receptor 2A,Soluble cytochrome b562, ... | | Authors: | Chen, Z, Fan, L, Wang, S. | | Deposit date: | 2023-06-21 | | Release date: | 2024-02-28 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Flexible scaffold-based cheminformatics approach for polypharmacological drug design.
Cell, 187, 2024
|
|
8JT6
 
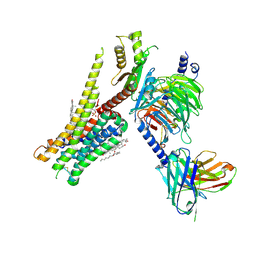 | | 5-HT1A-Gi in complex with compound (R)-IHCH-7179 | | Descriptor: | 1-(4-fluorophenyl)-4-[(7R)-2,5,11-triazatetracyclo[7.6.1.0^2,7.0^12,16]hexadeca-1(15),9,12(16),13-tetraen-5-yl]butan-1-one, CHOLESTEROL, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, ... | | Authors: | Chen, Z, Xu, P, Huang, S, Xu, H.E, Wang, S. | | Deposit date: | 2023-06-21 | | Release date: | 2024-02-28 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Flexible scaffold-based cheminformatics approach for polypharmacological drug design.
Cell, 187, 2024
|
|
5VF3
 
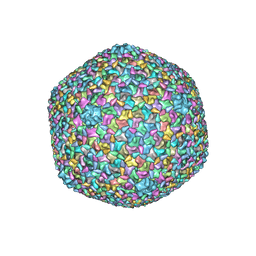 | | Bacteriophage T4 isometric capsid | | Descriptor: | Capsid vertex protein gp24, Highly immunogenic outer capsid protein, Major capsid protein, ... | | Authors: | Chen, Z, Sun, L, Zhang, Z, Fokine, A, Padilla-Sanchez, V, Hanein, D, Jiang, W, Rossmann, M.G, Rao, V.B. | | Deposit date: | 2017-04-06 | | Release date: | 2017-09-13 | | Last modified: | 2024-03-13 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Cryo-EM structure of the bacteriophage T4 isometric head at 3.3- angstrom resolution and its relevance to the assembly of icosahedral viruses.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
8EOI
 
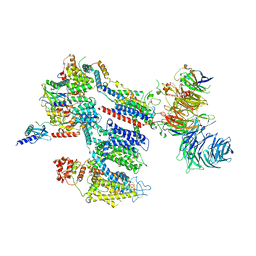 | | Structure of a human EMC:human Cav1.2 channel complex in GDN detergent | | Descriptor: | (3beta,14beta,17beta,25R)-3-[4-methoxy-3-(methoxymethyl)butoxy]spirost-5-en, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Chen, Z, Mondal, A, Abderemane-Ali, F, Minor, D.L. | | Deposit date: | 2022-10-03 | | Release date: | 2023-05-24 | | Last modified: | 2023-07-26 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | EMC chaperone-Ca V structure reveals an ion channel assembly intermediate.
Nature, 619, 2023
|
|
8EOG
 
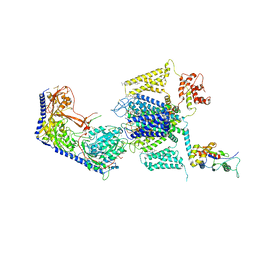 | | Structure of the human L-type voltage-gated calcium channel Cav1.2 complexed with L-leucine | | Descriptor: | (2R)-3-{[(R)-(2-aminoethoxy)(hydroxy)phosphoryl]oxy}-2-(dodecanoyloxy)propyl dodecanoate, (2R)-3-{[(R)-(2-aminoethoxy)(hydroxy)phosphoryl]oxy}-2-(dodecanoyloxy)propyl heptadecanoate, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Chen, Z, Mondal, A, Abderemane-Ali, F, Minor, D.L. | | Deposit date: | 2022-10-03 | | Release date: | 2023-05-24 | | Last modified: | 2023-07-26 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | EMC chaperone-Ca V structure reveals an ion channel assembly intermediate.
Nature, 619, 2023
|
|
8FD7
 
 | | Structure of the human L-type voltage-gated calcium channel Cav1.2 complexed with gabapentin | | Descriptor: | (2R)-3-{[(R)-(2-aminoethoxy)(hydroxy)phosphoryl]oxy}-2-(dodecanoyloxy)propyl heptadecanoate, (2S)-3-{[(R)-(2-aminoethoxy)(hydroxy)phosphoryl]oxy}-2-(dodecanoyloxy)propyl dodecanoate, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Chen, Z, Mondal, A, Minor, D.L. | | Deposit date: | 2022-12-02 | | Release date: | 2023-03-22 | | Last modified: | 2023-07-05 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structural basis for Ca V alpha 2 delta :gabapentin binding.
Nat.Struct.Mol.Biol., 30, 2023
|
|
