1AOR
 
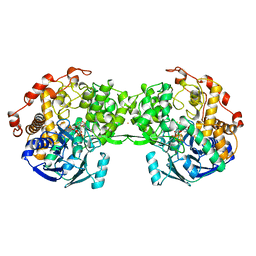 | | STRUCTURE OF A HYPERTHERMOPHILIC TUNGSTOPTERIN ENZYME, ALDEHYDE FERREDOXIN OXIDOREDUCTASE | | Descriptor: | ALDEHYDE FERREDOXIN OXIDOREDUCTASE, FE (III) ION, IRON/SULFUR CLUSTER, ... | | Authors: | Chan, M.K, Mukund, S, Kletzin, A, Adams, M.W.W, Rees, D.C. | | Deposit date: | 1995-02-13 | | Release date: | 1995-04-20 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of a hyperthermophilic tungstopterin enzyme, aldehyde ferredoxin oxidoreductase.
Science, 267, 1995
|
|
1DFF
 
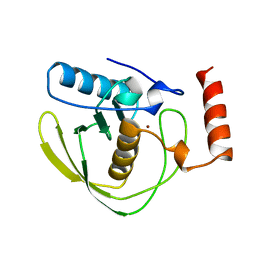 | | PEPTIDE DEFORMYLASE | | Descriptor: | PEPTIDE DEFORMYLASE, ZINC ION | | Authors: | Chan, M.K, Gong, W, Rajagopalan, P.T.R, Hao, B, Tsai, C.M, Pei, D. | | Deposit date: | 1997-08-19 | | Release date: | 1998-09-02 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.88 Å) | | Cite: | Crystal structure of the Escherichia coli peptide deformylase.
Biochemistry, 36, 1997
|
|
1BSJ
 
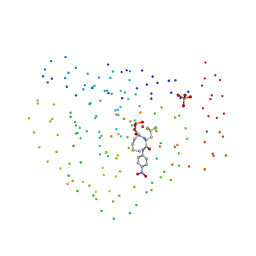 | | COBALT DEFORMYLASE INHIBITOR COMPLEX FROM E.COLI | | Descriptor: | (S)-2-(PHOSPHONOXY)CAPROYL-L-LEUCYL-P-NITROANILIDE, COBALT (II) ION, PHOSPHATE ION, ... | | Authors: | Hao, B, Gong, W, Rajagopalan, P.T, Hu, Y, Pei, D, Chan, M.K. | | Deposit date: | 1998-08-28 | | Release date: | 2000-04-15 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural basis for the design of antibiotics targeting peptide deformylase.
Biochemistry, 38, 1999
|
|
1BSK
 
 | | ZINC DEFORMYLASE INHIBITOR COMPLEX FROM E.COLI | | Descriptor: | (S)-2-(PHOSPHONOXY)CAPROYL-L-LEUCYL-P-NITROANILIDE, PHOSPHATE ION, PROTEIN (PEPTIDE DEFORMYLASE), ... | | Authors: | Hao, B, Gong, W, Rajagopalan, P.T, Hu, Y, Pei, D, Chan, M.K. | | Deposit date: | 1998-08-28 | | Release date: | 2000-04-15 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural basis for the design of antibiotics targeting peptide deformylase.
Biochemistry, 38, 1999
|
|
7XCL
 
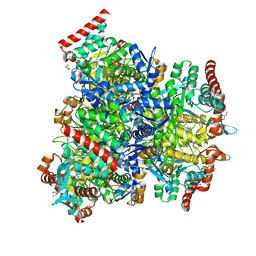 | |
7XCM
 
 | | Crystal structure of sulfite MttB structure at 3.2 A resolution | | Descriptor: | 3-METHYL-5-SULFO-PYRROLIDINE-2-CARBOXYLIC ACID, GLYCEROL, SODIUM ION, ... | | Authors: | Li, J, Chan, M.K. | | Deposit date: | 2022-03-24 | | Release date: | 2023-01-18 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Insights into pyrrolysine function from structures of a trimethylamine methyltransferase and its corrinoid protein complex.
Commun Biol, 6, 2023
|
|
7XCN
 
 | | Crystal structure of the MttB-MttC complex at 2.7 A resolution | | Descriptor: | 5-HYDROXYBENZIMIDAZOLYLCOBAMIDE, GLYCEROL, Trimethylamine methyltransferase, ... | | Authors: | Li, J, Chan, M.K. | | Deposit date: | 2022-03-24 | | Release date: | 2023-01-18 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Insights into pyrrolysine function from structures of a trimethylamine methyltransferase and its corrinoid protein complex.
Commun Biol, 6, 2023
|
|
8CXC
 
 | | Novel Anti-Mesothelin Antibodies Enable Crystallography of the Intact Mesothelin Ectodo- main and Engineering of Potent, T cell-engaging Bispecific Therapeutics | | Descriptor: | 3F2 Antibody heavy chain, 3F2 Antibody light chain, Mesothelin, ... | | Authors: | Bandaranayake, A.D, Rupert, P.B, Lin, I, Pilat, K, Ruff, R.O, Friend, D.J, Chan, M.K, Clarke, M, Carter, J, Meshinchi, S, Mehlin, C, Olson, J.M, Strong, R.K, Correnti, C.E. | | Deposit date: | 2022-05-20 | | Release date: | 2023-06-07 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (4.31 Å) | | Cite: | Novel mesothelin antibodies enable crystallography of the intact mesothelin ectodomain and engineering of potent, T cell-engaging bispecific therapeutics
Front. Drug Discov., 3, 2023
|
|
8CZ8
 
 | | Novel Anti-Mesothelin Antibodies Enable Crystallography of the Intact Mesothelin Ectodo- main and Engineering of Potent, T cell-engaging Bispecific Therapeutics | | Descriptor: | Mesothelin, cleaved form, SULFATE ION, ... | | Authors: | Bandaranayake, A.D, Rupert, P.B, Lin, I, Pilat, K, Ruff, R.O, Friend, D.J, Chan, M.K, Clarke, M, Carter, J, Meshinchi, S, Mehlin, C, Olson, J.M, Strong, R.K, Correnti, C.E. | | Deposit date: | 2022-05-24 | | Release date: | 2023-06-07 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Novel mesothelin antibodies enable crystallography of the intact mesothelin ectodomain and engineering of potent, T cell-engaging bispecific therapeutics
Front. Drug Discov., 3, 2023
|
|
8CYH
 
 | | Novel Anti-Mesothelin Antibodies Enable Crystallography of the Intact Mesothelin Ectodo- main and Engineering of Potent, T cell-engaging Bispecific Therapeutics | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, A12 antibody heavy chain, A12 antibody light chain, ... | | Authors: | Bandaranayake, A.D, Rupert, P.B, Lin, I, Pilat, K, Ruff, R.O, Friend, D.J, Chan, M.K, Clarke, M, Carter, J, Meshinchi, S, Mehlin, C, Olson, J.M, Strong, R.K, Correnti, C.E. | | Deposit date: | 2022-05-23 | | Release date: | 2023-06-07 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.38 Å) | | Cite: | Novel mesothelin antibodies enable crystallography of the intact mesothelin ectodomain and engineering of potent, T cell-engaging bispecific therapeutics
Front. Drug Discov., 3, 2023
|
|
1DP8
 
 | |
1DP9
 
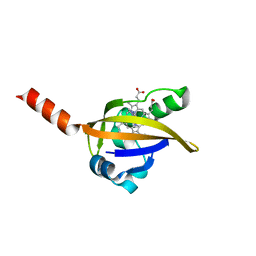 | | CRYSTAL STRUCTURE OF IMIDAZOLE-BOUND FIXL HEME DOMAIN | | Descriptor: | FIXL PROTEIN, IMIDAZOLE, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Gong, W, Hao, B, Chan, M.K. | | Deposit date: | 1999-12-24 | | Release date: | 2000-12-24 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | New mechanistic insights from structural studies of the oxygen-sensing domain of Bradyrhizobium japonicum FixL.
Biochemistry, 39, 2000
|
|
1DP6
 
 | | OXYGEN-BINDING COMPLEX OF FIXL HEME DOMAIN | | Descriptor: | FIXL PROTEIN, OXYGEN MOLECULE, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Gong, W, Hao, B, Chan, M.K. | | Deposit date: | 1999-12-23 | | Release date: | 2000-12-23 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | New mechanistic insights from structural studies of the oxygen-sensing domain of Bradyrhizobium japonicum FixL.
Biochemistry, 39, 2000
|
|
6JD9
 
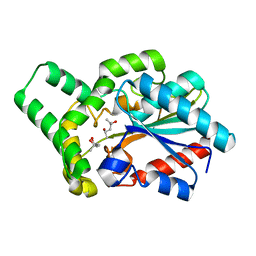 | | Proteus mirabilis lipase mutant - I118V/E130G | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, Alpha/beta hydrolase, CALCIUM ION | | Authors: | Heater, B.S, Chan, W.S, Chan, M.K. | | Deposit date: | 2019-01-31 | | Release date: | 2019-07-24 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Directed evolution of a genetically encoded immobilized lipase for the efficient production of biodiesel from waste cooking oil.
Biotechnol Biofuels, 12, 2019
|
|
1KA4
 
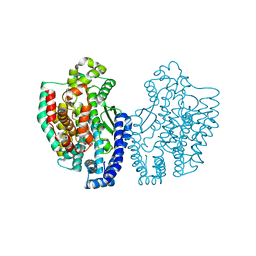 | | Structure of Pyrococcus furiosus carboxypeptidase Nat-Pb | | Descriptor: | LEAD (II) ION, M32 carboxypeptidase | | Authors: | Arndt, J.W, Hao, B, Ramakrishnan, V, Cheng, T, Chan, S.I, Chan, M.K. | | Deposit date: | 2001-10-31 | | Release date: | 2002-11-06 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal Structure of a Novel Carboxypeptidase from the Hyperthermophilic Archaeon Pyrococcus furiosus
Structure, 10, 2002
|
|
1K9X
 
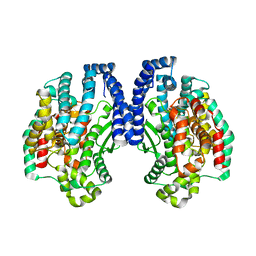 | | Structure of Pyrococcus furiosus carboxypeptidase Apo-Yb | | Descriptor: | M32 carboxypeptidase | | Authors: | Arndt, J.W, Hao, B, Ramakrishnan, V, Cheng, T, Chan, S.I, Chan, M.K. | | Deposit date: | 2001-10-31 | | Release date: | 2002-11-06 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structure of a Novel Carboxypeptidase from the Hyperthermophilic Archaeon Pyrococcus furiosus
Structure, 10, 2002
|
|
1KA2
 
 | | Structure of Pyrococcus furiosus Carboxypeptidase Apo-Mg | | Descriptor: | M32 carboxypeptidase, MAGNESIUM ION | | Authors: | Arndt, J.W, Hao, B, Ramakrishnan, V, Cheng, T, Chan, S.I, Chan, M.K. | | Deposit date: | 2001-10-31 | | Release date: | 2002-11-06 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal Structure of a Novel Carboxypeptidase from the Hyperthermophilic Archaeon Pyrococcus furiosus
Structure, 10, 2002
|
|
7EAR
 
 | | A positively charged mutant Cry3Aa endotoxin | | Descriptor: | Crystaline entomocidal protoxin | | Authors: | Yang, Z, Lee, M.M, Chan, M.K. | | Deposit date: | 2021-03-08 | | Release date: | 2021-04-28 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Efficient intracellular delivery of p53 protein by engineered protein crystals restores tumor suppressing function in vivo.
Biomaterials, 271, 2021
|
|
3HQ2
 
 | | BsuCP Crystal Structure | | Descriptor: | Bacillus subtilis M32 carboxypeptidase, CHLORIDE ION, FLUORIDE ION, ... | | Authors: | Lee, M.M, Isaza, C.E, White, J.D, Chen, R.P.-Y, Liang, G.F.-C, He, H.T.-F, Chan, S.I, Chan, M.K. | | Deposit date: | 2009-06-05 | | Release date: | 2009-06-30 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Insight into the substrate length restriction of M32 carboxypeptidases: Characterization of two distinct subfamilies.
Proteins, 77, 2009
|
|
8HHE
 
 | |
3HOA
 
 | |
3B9Z
 
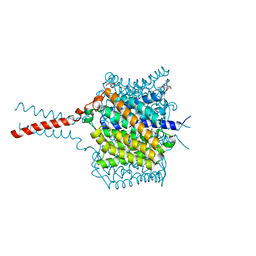 | | Crystal structure of the Nitrosomonas europaea Rh protein complexed with carbon dioxide | | Descriptor: | Ammonium transporter family Rh-like protein, CARBON DIOXIDE, UNKNOWN LIGAND, ... | | Authors: | Li, X, Jayachandran, S, Nguyen, H.-H.T, Chan, M.K. | | Deposit date: | 2007-11-07 | | Release date: | 2007-12-18 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structure of the Nitrosomonas europaea Rh protein.
Proc.Natl.Acad.Sci.Usa, 104, 2007
|
|
3B9Y
 
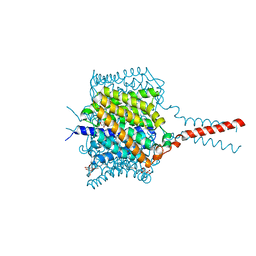 | | Crystal structure of the Nitrosomonas europaea Rh protein | | Descriptor: | Ammonium transporter family Rh-like protein, UNKNOWN LIGAND, octyl beta-D-glucopyranoside | | Authors: | Li, X, Jayachandran, S, Nguyen, H.-H.T, Chan, M.K. | | Deposit date: | 2007-11-07 | | Release date: | 2007-12-18 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structure of the Nitrosomonas europaea Rh protein.
Proc.Natl.Acad.Sci.Usa, 104, 2007
|
|
1DRM
 
 | | CRYSTAL STRUCTURE OF THE LIGAND FREE BJFIXL HEME DOMAIN | | Descriptor: | PROTOPORPHYRIN IX CONTAINING FE, SENSOR PROTEIN FIXL | | Authors: | Gong, W, Hao, B, Mansy, S.S, Gonzalez, G, Gilles-Gonzalez, M.A, Chan, M.K. | | Deposit date: | 2000-01-06 | | Release date: | 2000-01-24 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structure of a biological oxygen sensor: a new mechanism for heme-driven signal transduction.
Proc.Natl.Acad.Sci.USA, 95, 1998
|
|
4RPF
 
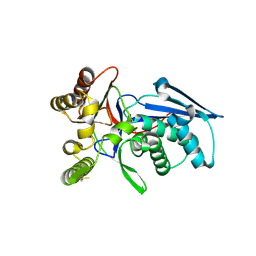 | | Crystal structure of homoserine kinase from Yersinia pestis Nepal516, NYSGRC target 032715 | | Descriptor: | CITRIC ACID, Homoserine kinase | | Authors: | Ptskovsky, Y, Bhosle, R, Toro, R, Hillerich, B, Gizzi, A, Garforth, S, Kar, A, Chan, M.K, Lafluer, J, Patel, H, Matikainen, B, Chamala, S, Lim, S, Celikgil, A, Villegas, G, Evans, B, Love, J, Fiser, A, Seidel, R, Bonanno, J.B, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2014-10-30 | | Release date: | 2014-11-12 | | Last modified: | 2018-01-24 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal Structure of Homoserine Kinase from Yersinia Pestis Nepal516, Nysgrc Target 032715
To be Published
|
|
