3L2C
 
 | | Crystal Structure of the DNA Binding Domain of FOXO4 Bound to DNA | | Descriptor: | FOXO consensus binding sequence, minus strand, plus strand, ... | | Authors: | Boura, E, Silhan, J, Sulc, M, Brynda, J, Obsilova, V, Obsil, T. | | Deposit date: | 2009-12-15 | | Release date: | 2009-12-29 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.868 Å) | | Cite: | Structure of the human FOXO4-DBD-DNA complex at 1.9 A resolution reveals new details of FOXO binding to the DNA
Acta Crystallogr.,Sect.D, 66, 2010
|
|
8BXK
 
 | | Ntaya virus methyltransferase | | Descriptor: | Ntaya virus methyltransferase, SINEFUNGIN | | Authors: | Boura, E, Krafcikova, P. | | Deposit date: | 2022-12-09 | | Release date: | 2023-01-18 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Ntaya virus methyltransferase
To Be Published
|
|
5NAS
 
 | |
5LX2
 
 | |
8PQO
 
 | |
3TOW
 
 | |
6YWA
 
 | |
6YWB
 
 | |
7A90
 
 | | WT STING in complex with 3',3'-c-di[2'FdAM(PS)] | | Descriptor: | 9-[(1~{R},3~{R},6~{R},8~{R},9~{R},10~{R},12~{R},15~{R},17~{R},18~{R})-17-(6-aminopurin-9-yl)-9,18-bis(fluoranyl)-3,12-bis(oxidanylidene)-3,12-bis(sulfanyl)-2,4,7,11,13,16-hexaoxa-3$l^{5},12$l^{5}-diphosphatricyclo[13.3.0.0^{6,10}]octadecan-8-yl]purin-6-amine, Stimulator of interferon protein | | Authors: | Boura, E, Smola, M. | | Deposit date: | 2020-09-01 | | Release date: | 2021-10-06 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (3.185 Å) | | Cite: | WT STING in complex with 3',3'-c-di[2'FdAM(PS)]
To Be Published
|
|
8CQH
 
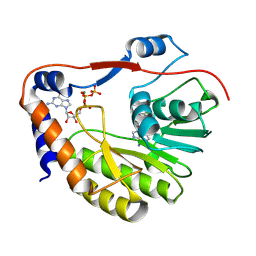 | | Ntaya virus methyltransferase in complex with GTP and SAH | | Descriptor: | GUANOSINE-5'-TRIPHOSPHATE, Genome polyprotein, MAGNESIUM ION, ... | | Authors: | Boura, E, Krejcova, K. | | Deposit date: | 2023-03-06 | | Release date: | 2024-03-13 | | Last modified: | 2024-09-04 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural and functional insights in flavivirus NS5 proteins gained by the structure of Ntaya virus polymerase and methyltransferase.
Structure, 32, 2024
|
|
4K7H
 
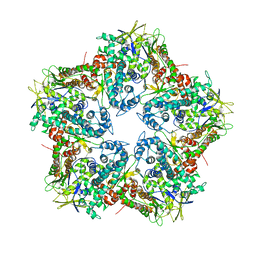 | | Major capsid protein P1 of the Pseudomonas phage phi6 | | Descriptor: | Major inner protein P1 | | Authors: | Boura, E, Nemecek, D, Plevka, P, Steven, C.A, Hurley, J.H. | | Deposit date: | 2013-04-17 | | Release date: | 2013-08-14 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3.5964 Å) | | Cite: | Subunit Folds and Maturation Pathway of a dsRNA Virus Capsid.
Structure, 21, 2013
|
|
5I14
 
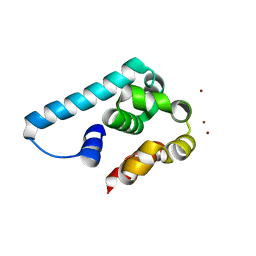 | | Truncated and mutated T4 lysozyme | | Descriptor: | NICKEL (II) ION, mutated and truncated T4 lysozyme | | Authors: | Klima, M, Boura, E. | | Deposit date: | 2016-02-05 | | Release date: | 2016-02-17 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.745 Å) | | Cite: | Metal ions-binding T4 lysozyme as an intramolecular protein purification tag compatible with X-ray crystallography.
Protein Sci., 26, 2017
|
|
2OJ4
 
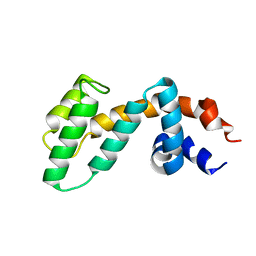 | | Crystal structure of RGS3 RGS domain | | Descriptor: | Regulator of G-protein signaling 3 | | Authors: | Boura, E, Obsil, T. | | Deposit date: | 2007-01-12 | | Release date: | 2007-01-30 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | 14-3-3 protein interacts with and affects the structure of RGS domain of regulator of G protein signaling 3 (RGS3).
J.Struct.Biol., 170, 2010
|
|
6QSN
 
 | | Crystal structure of Yellow fever virus polymerase NS5A | | Descriptor: | Genome polyprotein, S-ADENOSYL-L-HOMOCYSTEINE, SULFATE ION, ... | | Authors: | Boura, E, Dubankova, A. | | Deposit date: | 2019-02-21 | | Release date: | 2019-10-16 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (3.004 Å) | | Cite: | Structure of the yellow fever NS5 protein reveals conserved drug targets shared among flaviviruses.
Antiviral Res., 169, 2019
|
|
6S26
 
 | | Crystal structure of human wild type STING in complex with 2'-3'-cyclic-GMP-7-deaza-AMP | | Descriptor: | 2-azanyl-9-[(1~{R},6~{R},8~{R},9~{R},10~{S},15~{R},17~{R},18~{R})-8-(4-azanylpyrrolo[2,3-d]pyrimidin-7-yl)-3,9,12,18-tetrakis(oxidanyl)-3,12-bis(oxidanylidene)-2,4,7,11,13,16-hexaoxa-3$l^{5},12$l^{5}-diphosphatricyclo[13.2.1.0^{6,10}]octadecan-17-yl]-1~{H}-purin-6-one, Stimulator of interferon protein | | Authors: | Boura, E, Smola, M, Brynda, J. | | Deposit date: | 2019-06-20 | | Release date: | 2019-11-27 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Enzymatic Preparation of 2'-5',3'-5'-Cyclic Dinucleotides, Their Binding Properties to Stimulator of Interferon Genes Adaptor Protein, and Structure/Activity Correlations.
J.Med.Chem., 62, 2019
|
|
6Y99
 
 | | hSTING mutant R232K in complex with 2',3'-cGAMP | | Descriptor: | Stimulator of interferon genes protein, cGAMP | | Authors: | Boura, E, Smola, M. | | Deposit date: | 2020-03-06 | | Release date: | 2021-03-31 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.984 Å) | | Cite: | Ligand Strain and Its Conformational Complexity Is a Major Factor in the Binding of Cyclic Dinucleotides to STING Protein.
Angew.Chem.Int.Ed.Engl., 60, 2021
|
|
6YDB
 
 | | Human wtSTING in complex with 2',2'-difluoro-3',3'-c-di-GMP | | Descriptor: | 2-azanyl-9-[(1~{R},6~{R},8~{R},9~{R},10~{R},15~{R},17~{R},18~{R})-17-(2-azanyl-6-oxidanylidene-1~{H}-purin-9-yl)-9,18-bis(fluoranyl)-3,12-bis(oxidanyl)-3,12-bis(oxidanylidene)-2,4,7,11,13,16-hexaoxa-3$l^{5},12$l^{5}-diphosphatricyclo[13.3.0.0^{6,10}]octadecan-8-yl]-1~{H}-purin-6-one, Stimulator of interferon protein | | Authors: | Boura, E, Smola, M. | | Deposit date: | 2020-03-20 | | Release date: | 2021-03-31 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.801 Å) | | Cite: | Ligand Strain and Its Conformational Complexity Is a Major Factor in the Binding of Cyclic Dinucleotides to STING Protein.
Angew.Chem.Int.Ed.Engl., 60, 2021
|
|
6YDZ
 
 | | Human wtSTING in complex with 3',3'-cGAMP | | Descriptor: | 2-amino-9-[(2R,3R,3aS,5R,7aR,9R,10R,10aS,12R,14aR)-9-(6-amino-9H-purin-9-yl)-3,5,10,12-tetrahydroxy-5,12-dioxidooctahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8]tetraoxadiphosphacyclododecin-2-yl]-1,9-dihydro-6H-purin-6-one, Stimulator of interferon protein | | Authors: | Boura, E, Smola, M. | | Deposit date: | 2020-03-23 | | Release date: | 2021-03-31 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Ligand Strain and Its Conformational Complexity Is a Major Factor in the Binding of Cyclic Dinucleotides to STING Protein.
Angew.Chem.Int.Ed.Engl., 60, 2021
|
|
6YEA
 
 | | Human wtSTING in complex with 2',2'-difluoro-3',3'-cGAMP | | Descriptor: | 2',2'-difluoro-3',3'-cGAMP, Stimulator of interferon protein | | Authors: | Boura, E, Smola, M. | | Deposit date: | 2020-03-24 | | Release date: | 2021-03-31 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.805 Å) | | Cite: | Ligand Strain and Its Conformational Complexity Is a Major Factor in the Binding of Cyclic Dinucleotides to STING Protein.
Angew.Chem.Int.Ed.Engl., 60, 2021
|
|
6Z15
 
 | | Human wtSTING in complex with 3',3'-c-di-AMP | | Descriptor: | (2R,3R,3aS,5R,7aR,9R,10R,10aS,12R,14aR)-2,9-bis(6-amino-9H-purin-9-yl)octahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8 ]tetraoxadiphosphacyclododecine-3,5,10,12-tetrol 5,12-dioxide, Stimulator of interferon protein | | Authors: | Boura, E, Smola, M. | | Deposit date: | 2020-05-12 | | Release date: | 2021-04-21 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Ligand Strain and Its Conformational Complexity Is a Major Factor in the Binding of Cyclic Dinucleotides to STING Protein.
Angew.Chem.Int.Ed.Engl., 60, 2021
|
|
6Z0Z
 
 | | Human wtSTING in complex with 3',3'-c-(2'FdAMP-2'FdAMP) | | Descriptor: | 2'-fluoro-,3',3'-c-di-AMP, Stimulator of interferon protein | | Authors: | Boura, E, Smola, M. | | Deposit date: | 2020-05-11 | | Release date: | 2021-05-19 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.499 Å) | | Cite: | Ligand Strain and Its Conformational Complexity Is a Major Factor in the Binding of Cyclic Dinucleotides to STING Protein.
Angew.Chem.Int.Ed.Engl., 60, 2021
|
|
9FEH
 
 | | Crystal structure of SARS-CoV-2 nsp14 methyltransferase domain in complex with the STM957 inhibitor | | Descriptor: | Transcription factor ETV6,Guanine-N7 methyltransferase nsp14, ZINC ION, ~{N}-[[(2~{R},3~{S},4~{R},5~{R})-5-[4-azanyl-5-(2-pyridin-3-ylethynyl)pyrrolo[2,3-d]pyrimidin-7-yl]-3,4-bis(oxidanyl)oxolan-2-yl]methyl]-3-cyano-~{N}-ethyl-4-methoxy-benzenesulfonamide | | Authors: | Zilecka, E, Klima, M, Boura, E. | | Deposit date: | 2024-05-20 | | Release date: | 2024-08-28 | | Last modified: | 2024-09-04 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Structure of SARS-CoV-2 MTase nsp14 with the inhibitor STM957 reveals inhibition mechanism that is shared with a poxviral MTase VP39.
J Struct Biol X, 10, 2024
|
|
8QDJ
 
 | |
4PLA
 
 | |
5FBQ
 
 | | PI4KB in complex with Rab11 and the MI358 Inhibitor | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, Phosphatidylinositol 4-kinase beta,Phosphatidylinositol 4-kinase beta, Ras-related protein Rab-11A, ... | | Authors: | Chalupska, D, Mejdrova, I, Nencka, R, Boura, E. | | Deposit date: | 2015-12-14 | | Release date: | 2016-12-28 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.789 Å) | | Cite: | PI4KB in complex with Rab11 and the MI358 Inhibitor
To Be Published
|
|
