9IHS
 
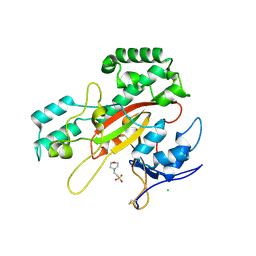 | | Microbial transglutaminase mutant - D3C/G283C | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Suzuki, M, Date, M, Kashiwagi, T, Takahashi, K, Nakamura, A, Tanokura, M, Suzuki, E, Yokoyama, K. | | Deposit date: | 2024-06-18 | | Release date: | 2024-09-18 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Random mutagenesis and disulfide bond formation improved thermostability in microbial transglutaminase.
Appl.Microbiol.Biotechnol., 108, 2024
|
|
1FIM
 
 | |
2JZ7
 
 | |
5HWA
 
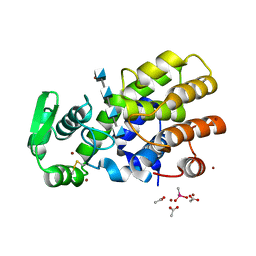 | | Crystal Structure of MH-K1 chitosanase in substrate-bound form | | Descriptor: | 2-amino-2-deoxy-beta-D-glucopyranose-(1-4)-2-amino-2-deoxy-beta-D-glucopyranose-(1-4)-2-amino-2-deoxy-beta-D-glucopyranose-(1-4)-2-amino-2-deoxy-beta-D-glucopyranose, ACETIC ACID, CACODYLATE ION, ... | | Authors: | Suzuki, M, Saito, A, Ando, A, Miki, K, Saito, J. | | Deposit date: | 2016-01-29 | | Release date: | 2017-02-08 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Crystal structure of the GH-46 subclass III chitosanase from Bacillus circulans MH-K1 in complex with chitotetraose
Biomed.Biochim.Acta, 1868, 2024
|
|
1F16
 
 | |
1PC2
 
 | |
1UIZ
 
 | | Crystal Structure Of Macrophage Migration Inhibitory Factor From Xenopus Laevis. | | Descriptor: | Macrophage Migration Inhibitory Factor | | Authors: | Suzuki, M, Takamura, Y, Maeno, M, Tochinai, S, Iyaguchi, D, Tanaka, I, Nishihira, J, Ishibashi, T. | | Deposit date: | 2003-07-24 | | Release date: | 2004-05-25 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Xenopus laevis Macrophage Migration Inhibitory Factor Is Essential for Axis Formation and Neural Development.
J.Biol.Chem., 279, 2004
|
|
1Y8M
 
 | |
6KTK
 
 | | Crystal structure of scyllo-inositol dehydrogenase R178A mutant, complexed with NADH and L-glucono-1,5-lactone, from Paracoccus laeviglucosivorans | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, L-glucono-1,5-lactone, Scyllo-inositol dehydrogenase with L-glucose dehydrogenase activity, ... | | Authors: | Suzuki, M, Koubara, K, Takenoya, M, Fukano, K, Ito, S, Sasaki, Y, Nakamura, A, Yajima, S. | | Deposit date: | 2019-08-28 | | Release date: | 2019-12-25 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Single amino acid mutation altered substrate specificity for L-glucose and inositol inscyllo-inositol dehydrogenase isolated fromParacoccus laeviglucosivorans.
Biosci.Biotechnol.Biochem., 84, 2020
|
|
6KTL
 
 | | Crystal structure of scyllo-inositol dehydrogenase R178A mutant, complexed with NAD and myo-inositol, from Paracoccus laeviglucosivorans | | Descriptor: | 1,2,3,4,5,6-HEXAHYDROXY-CYCLOHEXANE, ACETATE ION, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Suzuki, M, Koubara, K, Takenoya, M, Fukano, K, Ito, S, Sasaki, Y, Nakamura, A, Yajima, S. | | Deposit date: | 2019-08-28 | | Release date: | 2019-12-25 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Single amino acid mutation altered substrate specificity for L-glucose and inositol inscyllo-inositol dehydrogenase isolated fromParacoccus laeviglucosivorans.
Biosci.Biotechnol.Biochem., 84, 2020
|
|
6KTJ
 
 | | Crystal structure of scyllo-inositol dehydrogenase R178A mutant, apo-form, from Paracoccus laeviglucosivorans | | Descriptor: | ACETATE ION, Scyllo-inositol dehydrogenase with L-glucose dehydrogenase activity | | Authors: | Suzuki, M, Koubara, K, Takenoya, M, Fukano, K, Ito, S, Sasaki, Y, Nakamura, A, Yajima, S. | | Deposit date: | 2019-08-28 | | Release date: | 2019-12-25 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Single amino acid mutation altered substrate specificity for L-glucose and inositol inscyllo-inositol dehydrogenase isolated fromParacoccus laeviglucosivorans.
Biosci.Biotechnol.Biochem., 84, 2020
|
|
3VOT
 
 | | Crystal structure of L-amino acid ligase from Bacillus licheniformis | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Suzuki, M, Takahashi, Y, Noguchi, A, Arai, T, Yagasaki, M, Kino, K, Saito, J. | | Deposit date: | 2012-02-08 | | Release date: | 2012-11-07 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The structure of L-amino-acid ligase from Bacillus licheniformis
Acta Crystallogr.,Sect.D, 68, 2012
|
|
8WTR
 
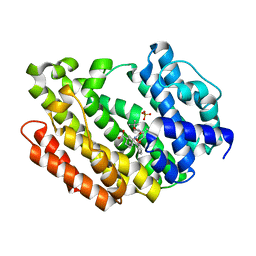 | | HUMAN SQUALENE SYNTHASE IN COMPLEX WITH (1S,3R)-3-[2-Chloro-5-(2,2-dimethyl-propyl)-13-(2-methoxy-phenyl)-6,8-dioxo-5,6,7,8,10,11-hexahydro-13H-12-oxa-5,9-diaza-benzocycloundecen-9-yl]-cyclohexanecarboxylic acid | | Descriptor: | (1~{S},3~{R})-3-[(10~{S})-13-chloranyl-2-(2,2-dimethylpropyl)-10-(2-methoxyphenyl)-3,5-bis(oxidanylidene)-9-oxa-2,6-diazabicyclo[9.4.0]pentadeca-1(15),11,13-trien-6-yl]cyclohexane-1-carboxylic acid, PHOSPHATE ION, Squalene synthase | | Authors: | Suzuki, M, Haginoya, N, Suzuki, M, Ishigai, Y, Terayama, K, Kanda, A, Sugita, K. | | Deposit date: | 2023-10-19 | | Release date: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Discovery of Novel 11-Membered Templates as Squalene Synthase Inhibitors.
J.Med.Chem., 67, 2024
|
|
8WTQ
 
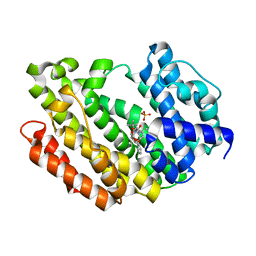 | | HUMAN SQUALENE SYNTHASE IN COMPLEX WITH {1-[2-Chloro-5-(2,2-dimethyl-propyl)-13-(2-methoxy-phenyl)-6-oxo-6,7,10,11-tetrahydro-5H,9H,13H-12-oxa-5,8-diaza-benzocycloundecene-8-carbonyl]-piperidin-4-yl}-acetic acid | | Descriptor: | 2-[1-[[(10~{S})-13-chloranyl-2-(2,2-dimethylpropyl)-10-(2-methoxyphenyl)-3-oxidanylidene-9-oxa-2,5-diazabicyclo[9.4.0]pentadeca-1(15),11,13-trien-5-yl]carbonyl]piperidin-4-yl]ethanoic acid, PHOSPHATE ION, Squalene synthase | | Authors: | Suzuki, M, Haginoya, N, Suzuki, M, Ishigai, Y, Terayama, K, Kanda, A, Sugita, K. | | Deposit date: | 2023-10-19 | | Release date: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Discovery of Novel 11-Membered Templates as Squalene Synthase Inhibitors.
J.Med.Chem., 67, 2024
|
|
7BTV
 
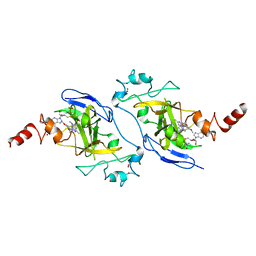 | | Crystal structure of EHMT2 SET domain in complex with compound 5. | | Descriptor: | Histone-lysine N-methyltransferase EHMT2, N~2~-{4-methoxy-3-[3-(pyrrolidin-1-yl)propoxy]phenyl}-N~4~,6-dimethylpyrimidine-2,4-diamine, S-ADENOSYLMETHIONINE, ... | | Authors: | Suzuki, M, Mizuno, T, Katayama, K. | | Deposit date: | 2020-04-03 | | Release date: | 2020-11-11 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Discovery of novel histone lysine methyltransferase G9a/GLP (EHMT2/1) inhibitors: Design, synthesis, and structure-activity relationships of 2,4-diamino-6-methylpyrimidines.
Bioorg.Med.Chem.Lett., 30, 2020
|
|
2D1J
 
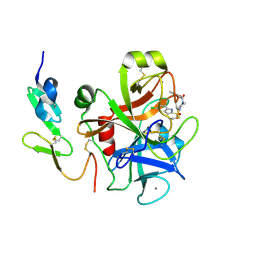 | | Factor Xa in complex with the inhibitor 2-[[4-[(5-chloroindol-2-yl)sulfonyl]piperazin-1-yl] carbonyl]thieno[3,2-b]pyridine n-oxide | | Descriptor: | 2-({4-[(5-CHLORO-1H-INDOL-2-YL)SULFONYL]PIPERAZIN-1-YL}CARBONYL)THIENO[3,2-B]PYRIDINE 4-OXIDE, CALCIUM ION, Coagulation factor X, ... | | Authors: | Suzuki, M. | | Deposit date: | 2005-08-24 | | Release date: | 2006-08-24 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Design, synthesis, and biological activity of non-basic compounds as factor Xa inhibitors: SAR study of S1 and aryl binding sites.
Bioorg.Med.Chem., 13, 2005
|
|
3H7G
 
 | | Apo-FR with AU ions | | Descriptor: | CADMIUM ION, Ferritin light chain, GLYCEROL, ... | | Authors: | Abe, M, Ueno, T, Abe, S, Suzuki, M, Goto, T, Toda, Y, Akita, T, Yamada, Y, Watanabe, Y. | | Deposit date: | 2009-04-27 | | Release date: | 2009-09-15 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Preparation and catalytic reaction of Au/Pd bimetallic nanoparticles in apo-ferritin
Chem.Commun.(Camb.), 32, 2009
|
|
1WU1
 
 | |
8GNE
 
 | | Crystal structure of human adenosine A2A receptor in complex with an insurmountable inverse agonist, KW-6356. | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 2-{2-[2-(2-{2-[2-(2-ETHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHOXY]-ETHOXY}-ETHANOL, Adenosine receptor A2a,Soluble cytochrome b562, ... | | Authors: | Suzuki, M, Saito, J, Miyagi, H, Yasunaga, M. | | Deposit date: | 2022-08-23 | | Release date: | 2023-03-22 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | In Vitro Pharmacological Profile of KW-6356, a Novel Adenosine A 2A Receptor Antagonist/Inverse Agonist.
Mol.Pharmacol., 103, 2023
|
|
8GNG
 
 | | Crystal structure of human adenosine A2A receptor in complex with istradefylline. | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 8-[(~{E})-2-(3,4-dimethoxyphenyl)ethenyl]-1,3-diethyl-7-methyl-purine-2,6-dione, Adenosine receptor A2a, ... | | Authors: | Suzuki, M, Saito, J, Miyagi, H, Yasunaga, M. | | Deposit date: | 2022-08-23 | | Release date: | 2023-03-22 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | In Vitro Pharmacological Profile of KW-6356, a Novel Adenosine A 2A Receptor Antagonist/Inverse Agonist.
Mol.Pharmacol., 103, 2023
|
|
3IIT
 
 | | Factor XA in complex with a cis-1,2-diaminocyclohexane derivative | | Descriptor: | 7-chloro-N-[(1S,2R,4S)-4-(dimethylcarbamoyl)-2-{[(5-methyl-5,6-dihydro-4H-pyrrolo[3,4-d][1,3]thiazol-2-yl)carbonyl]amino}cyclohexyl]isoquinoline-3-carboxamide, Activated factor Xa heavy chain, CALCIUM ION, ... | | Authors: | Suzuki, M. | | Deposit date: | 2009-08-03 | | Release date: | 2010-08-04 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Design, synthesis, and SAR of cis-1,2-diaminocyclohexane derivatives as potent factor Xa inhibitors. Part II: exploration of 6-6 fused rings as alternative S1 moieties.
Bioorg.Med.Chem., 17, 2009
|
|
3Q2Z
 
 | | Human Squalene synthase in complex with N-[(3R,5S)-7-Chloro-5-(2,3-dimethoxyphenyl)-1-neopentyl-2-oxo-1,2,3,5-tetrahydro-4,1-benzoxazepine-3-acetyl]-L-aspartic acid | | Descriptor: | N-{[(3R,5S)-7-chloro-5-(2,3-dimethoxyphenyl)-1-(2,2-dimethylpropyl)-2-oxo-1,2,3,5-tetrahydro-4,1-benzoxazepin-3-yl]acetyl}-L-aspartic acid, PHOSPHATE ION, Squalene synthase | | Authors: | Suzuki, M, Shimizu, H, Katakura, S, Yamazaki, K, Higashihashi, N, Ichikawa, M, Yokomizo, A, Itoh, M, Sugita, K, Usui, H. | | Deposit date: | 2010-12-21 | | Release date: | 2011-12-21 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Discovery of a new 2-aminobenzhydrol template for highly potent squalene synthase inhibitors
Bioorg.Med.Chem., 19, 2011
|
|
3Q30
 
 | | Human Squalene synthase in complex with (2R,3R)-2-Carboxymethoxy-3-[5-(2-naphthalenyl)pentyl]aminocarbonyl-3-[5-(2-naphthalenyl)pentyloxy]propionic acid | | Descriptor: | (2R,3R)-2-(carboxymethoxy)-4-{[5-(naphthalen-2-yl)pentyl]amino}-3-{[5-(naphthalen-2-yl)pentyl]oxy}-4-oxobutanoic acid, MAGNESIUM ION, PHOSPHATE ION, ... | | Authors: | Suzuki, M, Shimizu, H, Katakura, S, Yamazaki, K, Higashihashi, N, Ichikawa, M, Yokomizo, A, Itoh, M, Sugita, K, Usui, H. | | Deposit date: | 2010-12-21 | | Release date: | 2011-12-21 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Discovery of a new 2-aminobenzhydrol template for highly potent squalene synthase inhibitors
Bioorg.Med.Chem., 19, 2011
|
|
3Q3K
 
 | | Factor Xa in complex with a phenylenediamine derivative | | Descriptor: | Activated factor Xa heavy chain, CALCIUM ION, Factor X light chain, ... | | Authors: | Suzuki, M, Imai, E. | | Deposit date: | 2010-12-22 | | Release date: | 2011-12-28 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Design, synthesis and SAR of novel ethylenediamine and phenylenediamine derivatives as factor Xa inhibitors.
Bioorg.Med.Chem.Lett., 21, 2011
|
|
4QBJ
 
 | | Crystal structure of N-myristoyl transferase from Aspergillus fumigatus complexed with a synthetic inhibitor | | Descriptor: | 3-[[3-methyl-2-[[2,3,4-tris(fluoranyl)phenoxy]methyl]-1-benzofuran-4-yl]oxy]-N-(pyridin-3-ylmethyl)propan-1-amine, Glycylpeptide N-tetradecanoyltransferase, S-(2-OXO)PENTADECYLCOA, ... | | Authors: | Suzuki, M, Shimada, T. | | Deposit date: | 2014-05-08 | | Release date: | 2015-04-01 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure of N-myristoyltransferase from Aspergillus fumigatus
Acta Crystallogr.,Sect.D, 71, 2015
|
|
