6Y9A
 
 | |
6Y90
 
 | |
6Y97
 
 | |
6Y92
 
 | |
5N74
 
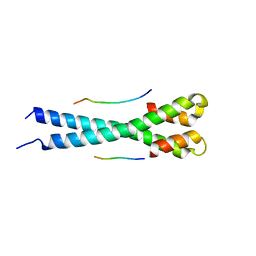 | | Microtubule end binding protein complex | | Descriptor: | Karyogamy protein KAR9, Microtubule-associated protein RP/EB family member 1 | | Authors: | Kumar, A, Steinmetz, M. | | Deposit date: | 2017-02-18 | | Release date: | 2017-06-14 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Short Linear Sequence Motif LxxPTPh Targets Diverse Proteins to Growing Microtubule Ends.
Structure, 25, 2017
|
|
5OAT
 
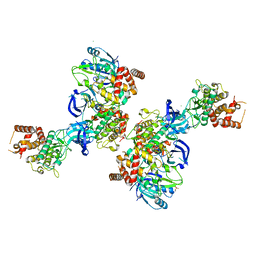 | | PINK1 structure | | Descriptor: | MAGNESIUM ION, Serine/threonine-protein kinase PINK1, mitochondrial-like Protein | | Authors: | Kumar, A, Tamjar, J, Woodroof, H.I, Raimi, O.G, Waddell, A.Y, Peggie, M, Muqit, M.M.K, van Aalten, D.M.F. | | Deposit date: | 2017-06-23 | | Release date: | 2017-10-11 | | Last modified: | 2017-10-18 | | Method: | X-RAY DIFFRACTION (2.78 Å) | | Cite: | Structure of PINK1 and mechanisms of Parkinson's disease associated mutations.
Elife, 6, 2017
|
|
5N2W
 
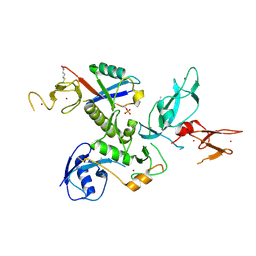 | | WT-Parkin and pUB complex | | Descriptor: | CHLORIDE ION, E3 ubiquitin-protein ligase parkin,E3 ubiquitin-protein ligase parkin, Polyubiquitin-B, ... | | Authors: | Kumar, A, Chaugule, V.K, Johnson, C, Toth, R, Sundaramoorthy, R, Knebel, A, Walden, H. | | Deposit date: | 2017-02-08 | | Release date: | 2017-04-19 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.68 Å) | | Cite: | Parkin-phosphoubiquitin complex reveals cryptic ubiquitin-binding site required for RBR ligase activity.
Nat. Struct. Mol. Biol., 24, 2017
|
|
5N38
 
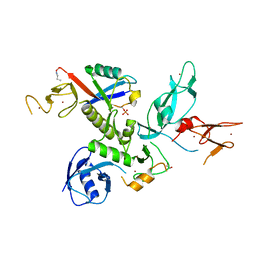 | | S65DParkin and pUB complex | | Descriptor: | CHLORIDE ION, DI(HYDROXYETHYL)ETHER, E3 ubiquitin-protein ligase parkin,E3 ubiquitin-protein ligase parkin, ... | | Authors: | Kumar, A, Chaugule, V.K, Johnson, C, Toth, R, Sundaramoorthy, R, Knebel, A, Walden, H. | | Deposit date: | 2017-02-08 | | Release date: | 2017-04-19 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Parkin-phosphoubiquitin complex reveals cryptic ubiquitin-binding site required for RBR ligase activity.
Nat. Struct. Mol. Biol., 24, 2017
|
|
3KJZ
 
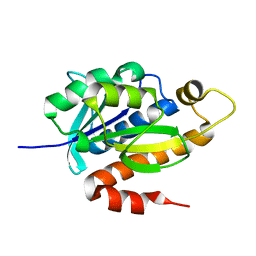 | | Crystal structure of native peptidyl-tRNA hydrolase from Mycobacterium smegmatis | | Descriptor: | Peptidyl-tRNA hydrolase | | Authors: | Kumar, A, Singh, N, Yadav, R, Prem Kumar, R, Sharma, S, Arora, A, Singh, T.P. | | Deposit date: | 2009-11-04 | | Release date: | 2010-08-18 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of peptidyl-tRNA hydrolase from mycobacterium smegmatis reveals novel features related to enzyme dynamics.
Int J Biochem Mol Biol, 3, 2012
|
|
7CXD
 
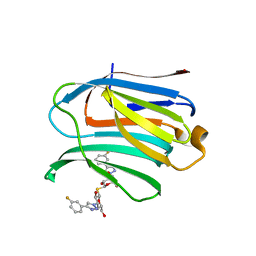 | | Xray structure of rat Galectin-3 CRD in complex with TD-139 belonging to P121 space group | | Descriptor: | 3-deoxy-3-[4-(3-fluorophenyl)-1H-1,2,3-triazol-1-yl]-beta-D-galactopyranosyl 3-deoxy-3-[4-(3-fluorophenyl)-1H-1,2,3-triazol-1-yl]-1-thio-beta-D-galactopyranoside, BROMIDE ION, Galectin-3 | | Authors: | Kumar, A. | | Deposit date: | 2020-09-01 | | Release date: | 2021-09-01 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | Molecular mechanism of interspecies differences in the binding affinity of TD139 to Galectin-3.
Glycobiology, 31, 2021
|
|
7CXA
 
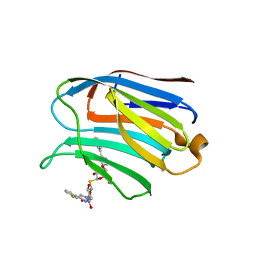 | | Structure of human Galectin-3 CRD in complex with TD-139 belonging to P31 space group. | | Descriptor: | 3-deoxy-3-[4-(3-fluorophenyl)-1H-1,2,3-triazol-1-yl]-beta-D-galactopyranosyl 3-deoxy-3-[4-(3-fluorophenyl)-1H-1,2,3-triazol-1-yl]-1-thio-beta-D-galactopyranoside, CHLORIDE ION, Galectin-3 | | Authors: | Kumar, A. | | Deposit date: | 2020-09-01 | | Release date: | 2021-09-01 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Molecular mechanism of interspecies differences in the binding affinity of TD139 to Galectin-3.
Glycobiology, 31, 2021
|
|
7CXC
 
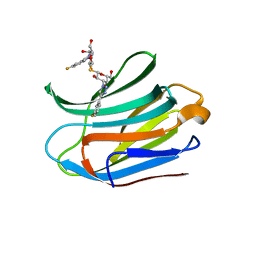 | |
7CXB
 
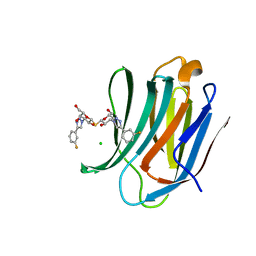 | | Structure of mouse Galectin-3 CRD in complex with TD-139 belonging to P6522 space group. | | Descriptor: | 3-deoxy-3-[4-(3-fluorophenyl)-1H-1,2,3-triazol-1-yl]-beta-D-galactopyranosyl 3-deoxy-3-[4-(3-fluorophenyl)-1H-1,2,3-triazol-1-yl]-1-thio-beta-D-galactopyranoside, CHLORIDE ION, Galectin-3 | | Authors: | Kumar, A. | | Deposit date: | 2020-09-01 | | Release date: | 2021-09-01 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.46 Å) | | Cite: | Molecular mechanism of interspecies differences in the binding affinity of TD139 to Galectin-3.
Glycobiology, 31, 2021
|
|
4LQ6
 
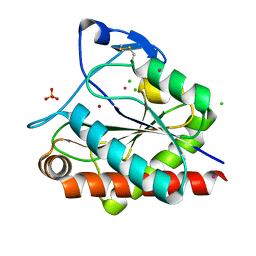 | | Crystal structure of Rv3717 reveals a novel amidase from M. tuberculosis | | Descriptor: | CHLORIDE ION, N-acetymuramyl-L-alanine amidase-related protein, PLATINUM (II) ION, ... | | Authors: | Kumar, A, Kumar, S, Kumar, D, Mishra, A, Dewangan, R.P, Shrivastava, P, Ramachandran, S, Taneja, B. | | Deposit date: | 2013-07-17 | | Release date: | 2013-12-04 | | Last modified: | 2014-01-15 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | The structure of Rv3717 reveals a novel amidase from Mycobacterium tuberculosis.
Acta Crystallogr.,Sect.D, 69, 2013
|
|
7M6S
 
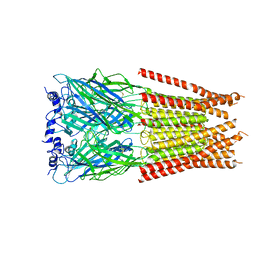 | |
7M6O
 
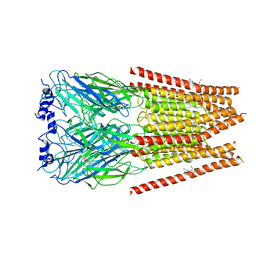 | | Full length alpha1 Glycine receptor in presence of 0.1mM Glycine and 32uM Tetrahydrocannabinol | | Descriptor: | (6aR,10aR)-6,6,9-trimethyl-3-pentyl-6a,7,8,10a-tetrahydro-6H-benzo[c]chromen-1-ol, 2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCINE, ... | | Authors: | Kumar, A, Chakrapani, S. | | Deposit date: | 2021-03-26 | | Release date: | 2022-08-03 | | Last modified: | 2022-09-07 | | Method: | ELECTRON MICROSCOPY (2.84 Å) | | Cite: | Structural basis for cannabinoid-induced potentiation of alpha1-glycine receptors in lipid nanodiscs.
Nat Commun, 13, 2022
|
|
7M6P
 
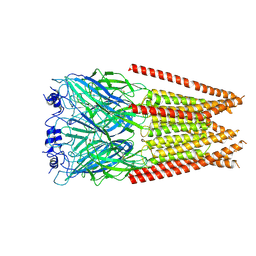 | | Full length alpha1 Glycine receptor in presence of 1mM Glycine | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCINE, Glycine receptor subunit alphaZ1 | | Authors: | Kumar, A, Chakrapani, S. | | Deposit date: | 2021-03-26 | | Release date: | 2022-08-03 | | Last modified: | 2022-09-07 | | Method: | ELECTRON MICROSCOPY (3.28 Å) | | Cite: | Structural basis for cannabinoid-induced potentiation of alpha1-glycine receptors in lipid nanodiscs.
Nat Commun, 13, 2022
|
|
7M6Q
 
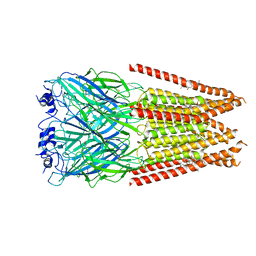 | | Full length alpha1 Glycine receptor in presence of 1mM Glycine and 32uM Tetrahydrocannabinol State 1 | | Descriptor: | (6aR,10aR)-6,6,9-trimethyl-3-pentyl-6a,7,8,10a-tetrahydro-6H-benzo[c]chromen-1-ol, 2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCINE, ... | | Authors: | Kumar, A, Chakrapani, S. | | Deposit date: | 2021-03-26 | | Release date: | 2022-08-03 | | Last modified: | 2022-09-07 | | Method: | ELECTRON MICROSCOPY (2.91 Å) | | Cite: | Structural basis for cannabinoid-induced potentiation of alpha1-glycine receptors in lipid nanodiscs.
Nat Commun, 13, 2022
|
|
7M6N
 
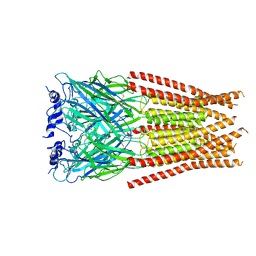 | |
7M6M
 
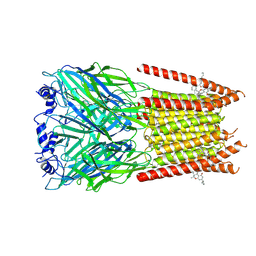 | | Full length alpha1 Glycine receptor in presence of 32uM Tetrahydrocannabinol | | Descriptor: | (6aR,10aR)-6,6,9-trimethyl-3-pentyl-6a,7,8,10a-tetrahydro-6H-benzo[c]chromen-1-ol, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Glycine receptor subunit alphaZ1 | | Authors: | Kumar, A, Chakrapani, S. | | Deposit date: | 2021-03-26 | | Release date: | 2022-08-03 | | Last modified: | 2022-09-07 | | Method: | ELECTRON MICROSCOPY (3.09 Å) | | Cite: | Structural basis for cannabinoid-induced potentiation of alpha1-glycine receptors in lipid nanodiscs.
Nat Commun, 13, 2022
|
|
7M6R
 
 | |
6JQV
 
 | | Crystal structure of Arabidopsis thaliana NRP2 | | Descriptor: | NAP1-related protein 2 | | Authors: | Kumar, A, Vasudevan, D. | | Deposit date: | 2019-04-01 | | Release date: | 2019-07-10 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.42 Å) | | Cite: | Structural Characterization ofArabidopsis thalianaNAP1-Related Protein 2 (AtNRP2) and Comparison with its Homolog AtNRP1.
Molecules, 24, 2019
|
|
7MWW
 
 | | Structure of hepatitis C virus envelope full-length glycoprotein 2 (eE2) from J6 genotype | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2A12 Fab Heavy chain, ... | | Authors: | Kumar, A, Hossain, R.A, Yost, S.A, Bu, W, Wang, Y, Dearborn, A.D, Grakoui, A, Cohen, J.I, Marcotrigiano, J. | | Deposit date: | 2021-05-17 | | Release date: | 2021-09-15 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.71 Å) | | Cite: | Structural insights into hepatitis C virus receptor binding and entry.
Nature, 598, 2021
|
|
7MWS
 
 | | Crystal structure of tamarin CD81 large extracellular loop | | Descriptor: | CD81 protein, GLYCEROL, TETRAETHYLENE GLYCOL | | Authors: | Kumar, A, Hossain, R.A, Yost, S.A, Bu, W, Wang, Y, Dearborn, A.D, Grakoui, A, Cohen, J.I, Marcotrigiano, J. | | Deposit date: | 2021-05-17 | | Release date: | 2021-09-15 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural insights into hepatitis C virus receptor binding and entry.
Nature, 598, 2021
|
|
7MWX
 
 | | Structure of the core ectodomain of the hepatitis C virus envelope glycoprotein 2 with tamarin CD81 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2A12 Fab Heavy Chain, ... | | Authors: | Kumar, A, Hossain, R.A, Yost, S.A, Bu, W, Wang, Y, Dearborn, A.D, Grakoui, A, Cohen, J.I, Marcotrigiano, J. | | Deposit date: | 2021-05-17 | | Release date: | 2021-09-15 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.32 Å) | | Cite: | Structural insights into hepatitis C virus receptor binding and entry.
Nature, 598, 2021
|
|
