7QA9
 
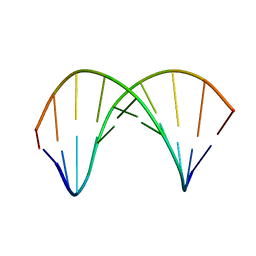 | | 10bp DNA/DNA duplex | | Descriptor: | DNA (5'-D(*CP*CP*AP*TP*TP*AP*TP*AP*GP*C)-3'), DNA (5'-D(*GP*CP*TP*AP*TP*AP*AP*TP*GP*G)-3') | | Authors: | Li, Q, Trajkovski, M, Fan, C, Chen, J, Zhou, Y, Lu, K, Li, H, Su, X, Xi, Z, Plavec, J, Zhou, C. | | Deposit date: | 2021-11-16 | | Release date: | 2022-11-16 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | 4'-SCF 3 -Labeling Constitutes a Sensitive 19 F NMR Probe for Characterization of Interactions in the Minor Groove of DNA.
Angew.Chem.Int.Ed.Engl., 61, 2022
|
|
2F7X
 
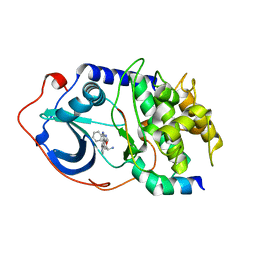 | | Protein Kinase A bound to (S)-2-(1H-Indol-3-yl)-1-[5-((E)-2-pyridin-4-yl-vinyl)-pyridin-3-yloxymethyl]-ethylamine | | Descriptor: | (1S)-2-(1H-INDOL-3-YL)-1-[({5-[(E)-2-PYRIDIN-4-YLVINYL]PYRIDIN-3-YL}OXY)METHYL]ETHYLAMINE, PKI, inhibitory peptide, ... | | Authors: | Li, Q, Li, T, Zhu, G.D, Gong, J, Claibone, A, Dalton, C, Luo, Y, Johnson, E.F, Shi, Y, Liu, X, Klinghofer, V, Bauch, J.L, Marsh, K.C, Bouska, J.J, Arries, S, De Jong, R, Oltersdorf, T, Stoll, V.S, Jakob, C.G, Rosenberg, S.H, Giranda, V.L. | | Deposit date: | 2005-12-01 | | Release date: | 2006-06-27 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Discovery of trans-3,4'-bispyridinylethylenes as potent and novel inhibitors of protein kinase B (PKB/Akt) for the treatment of cancer: Synthesis and biological evaluation.
Bioorg.Med.Chem.Lett., 16, 2006
|
|
2F7Z
 
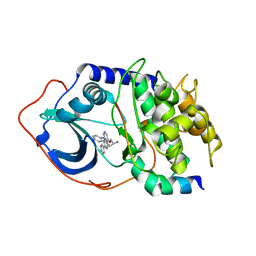 | | Protein Kinase A bound to (R)-1-(1H-Indol-3-ylmethyl)-2-(2-pyridin-4-yl-[1,7]naphtyridin-5-yloxy)-ehylamine | | Descriptor: | (1S)-1-(1H-INDOL-3-YLMETHYL)-2-(2-PYRIDIN-4-YL-[1,7]NAPHTYRIDIN-5-YLOXY)-EHYLAMINE, PKI, inhibitory peptide, ... | | Authors: | Li, Q, Woods, K.W, Thomas, S, Zhu, G.D, Packard, G, Fisher, J, Li, T, Gong, J, Dinges, J, Song, X, Abrams, J, Luo, Y, Johnson, E.F, Shi, Y, Liu, X, Klinghofer, V, Des Jong, R, Oltersdorf, T, Stoll, V.S, Jakob, C.G, Rosenberg, S.H, Giranda, V.L. | | Deposit date: | 2005-12-01 | | Release date: | 2006-06-27 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Synthesis and structure-activity relationship of 3,4'-bispyridinylethylenes: discovery of a potent 3-isoquinolinylpyridine inhibitor of protein kinase B (PKB/Akt) for the treatment of cancer.
Bioorg.Med.Chem.Lett., 16, 2006
|
|
5LGG
 
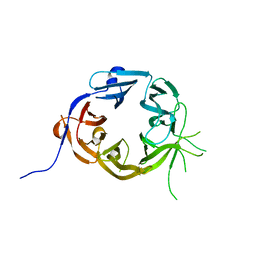 | | The N-terminal WD40 domain of Apc1 (Anaphase promoting complex subunit 1) | | Descriptor: | Anaphase-promoting complex subunit 1,Anaphase-promoting complex subunit 1,Anaphase-promoting complex subunit 1,Anaphase-promoting complex subunit 1 | | Authors: | Li, Q, Aibara, S, Barford, D. | | Deposit date: | 2016-07-07 | | Release date: | 2016-10-05 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | WD40 domain of Apc1 is critical for the coactivator-induced allosteric transition that stimulates APC/C catalytic activity.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
1X81
 
 | | Farnesyl transferase structure of Jansen compound | | Descriptor: | 6-[(S)-AMINO(4-CHLOROPHENYL)(1-METHYL-1H-IMIDAZOL-5-YL)METHYL]-4-(3-CHLOROPHENYL)-1-METHYLQUINOLIN-2(1H)-ONE, Protein farnesyltransferase beta subunit, Protein farnesyltransferase/geranylgeranyltransferase type I alpha subunit, ... | | Authors: | Li, Q, Claiborne, A, Li, T, Hasvold, L, Stoll, V.S, Muchmore, S, Jakob, C.G, Gu, W, Cohen, J, Hutchins, C, Frost, D, Rosenberg, S.H, Sham, H.L. | | Deposit date: | 2004-08-16 | | Release date: | 2004-12-21 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Design, synthesis, and activity of 4-quinolone and pyridone compounds as nonthiol-containing farnesyltransferase inhibitors.
Bioorg.Med.Chem.Lett., 14, 2004
|
|
4ZHS
 
 | |
2MV6
 
 | |
3PFT
 
 | | Crystal Structure of Untagged C54A Mutant Flavin Reductase (DszD) in Complex with FMN From Mycobacterium goodii | | Descriptor: | FLAVIN MONONUCLEOTIDE, Flavin reductase | | Authors: | Li, Q, Xu, P, Ma, C, Gu, L, Liu, X, Zhang, C, Li, N, Su, J, Li, B, Liu, S. | | Deposit date: | 2010-10-29 | | Release date: | 2011-11-02 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.601 Å) | | Cite: | The flavin reductase DSZD from a desulfurizing mycobacterium goodii strain: systemic manipulation and investigation based on the crystal structure
To be Published
|
|
4ZIC
 
 | |
8DJA
 
 | |
2MXB
 
 | |
6WK7
 
 | | Crystal Structure Analysis of a poly(thymine) DNA duplex | | Descriptor: | 1,3,5-triazine-2,4,6-triamine, DNA (5'-D(*TP*TP*TP*TP*TP*T)-3') | | Authors: | Li, Q, Zhao, J, Liu, L, Mandal, S, Rizzuto, F.J, He, H, Wei, S, Jonchhe, S, Sleiman, H.F, Mao, H, Mao, C. | | Deposit date: | 2020-04-15 | | Release date: | 2020-07-22 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.423 Å) | | Cite: | A poly(thymine)-melamine duplex for the assembly of DNA nanomaterials.
Nat Mater, 19, 2020
|
|
4FVK
 
 | | Structural and functional characterization of neuraminidase-like molecule N10 derived from bat influenza A virus | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Li, Q, Sun, X.M, Li, Z.X, Liu, Y, Vavricka, C.J, Qi, J.X, Gao, G.F. | | Deposit date: | 2012-06-29 | | Release date: | 2012-09-19 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.198 Å) | | Cite: | Structural and functional characterization of neuraminidase-like molecule N10 derived from bat influenza A virus
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
3NSS
 
 | | The 2009 pandemic H1N1 neuraminidase N1 lacks the 150-cavity in its active sites | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ACETATE ION, CALCIUM ION, ... | | Authors: | Li, Q, Qi, J.X, Zhang, W, Vavricka, C.J, Shi, Y, Gao, G.F. | | Deposit date: | 2010-07-02 | | Release date: | 2010-09-22 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.902 Å) | | Cite: | The 2009 pandemic H1N1 neuraminidase N1 lacks the 150-cavity in its active site
Nat.Struct.Mol.Biol., 17, 2010
|
|
1OR5
 
 | | SOLUTION STRUCTURE OF THE HOLO-FORM OF THE FRENOLICIN ACYL CARRIER PROTEIN, MINIMIZED MEAN STRUCTURE | | Descriptor: | acyl carrier protein | | Authors: | Li, Q, Khosla, C, Puglisi, J, Liu, C.W. | | Deposit date: | 2003-03-11 | | Release date: | 2003-07-01 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution Structure and Backbone Dynamics of the Holo Form of the Frenolicin Acyl Carrier Protein
Biochemistry, 42, 2003
|
|
6ITH
 
 | |
1ZCM
 
 | | Human calpain protease core inhibited by ZLLYCH2F | | Descriptor: | CALCIUM ION, Calpain 1, large [catalytic] subunit, ... | | Authors: | Li, Q, Hanzlik, R.P, Weaver, R.F, Schonbrunn, E. | | Deposit date: | 2005-04-12 | | Release date: | 2006-01-31 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Molecular mode of action of a covalently inhibiting peptidomimetic on the human calpain protease core
Biochemistry, 45, 2006
|
|
4OE1
 
 | | Crystal structure of the pentatricopeptide repeat protein PPR10 (C256S/C430S/C449S) in complex with an 18-nt PSAJ rna element | | Descriptor: | Chloroplast pentatricopeptide repeat protein 10, PHOSPHATE ION, psaJ RNA | | Authors: | Li, Q, Yan, C, Wu, J, Yin, P, Yan, N. | | Deposit date: | 2014-01-11 | | Release date: | 2014-09-24 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Examination of the dimerization states of the single-stranded RNA recognition protein pentatricopeptide repeat 10 (PPR10).
J.Biol.Chem., 289, 2014
|
|
4RFR
 
 | | Complex structure of AlkB/rhein | | Descriptor: | 4,5-dihydroxy-9,10-dioxo-9,10-dihydroanthracene-2-carboxylic acid, Alpha-ketoglutarate-dependent dioxygenase AlkB, MANGANESE (II) ION | | Authors: | Li, Q, Huang, Y, Li, J.F, Yang, C.-G. | | Deposit date: | 2014-09-27 | | Release date: | 2016-04-06 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Rhein Inhibits AlkB Repair Enzymes and Sensitizes Cells to Methylated DNA Damage.
J.Biol.Chem., 291, 2016
|
|
2I1J
 
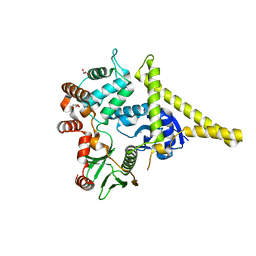 | | Moesin from Spodoptera frugiperda at 2.1 angstroms resolution | | Descriptor: | CHLORIDE ION, GLYCEROL, Moesin, ... | | Authors: | Li, Q, Nance, M.R, Tesmer, J.J.G. | | Deposit date: | 2006-08-14 | | Release date: | 2006-12-19 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Self-masking in an Intact ERM-merlin Protein: An Active Role for the Central alpha-Helical Domain.
J.Mol.Biol., 365, 2007
|
|
2MFR
 
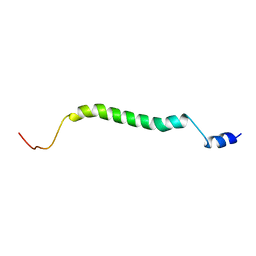 | |
7BJI
 
 | | Crystal structure of the Danio rerio centrosomal protein Cep135 coiled-coil fragment 64-190 | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, Centrosomal protein of 135 kDa, ... | | Authors: | Li, Q, Hatzopoulos, G, Iller, O, Vakonakis, I. | | Deposit date: | 2021-01-14 | | Release date: | 2021-01-27 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.58 Å) | | Cite: | Crystal structure of the Danio rerio centrosomal protein Cep135 coiled-coil fragment 64-190
To Be Published
|
|
2MHF
 
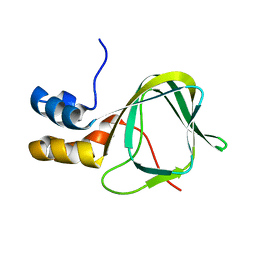 | |
7RL4
 
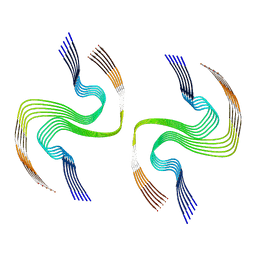 | |
7LC5
 
 | |
