3J9J
 
 | | Structure of the capsaicin receptor, TRPV1, determined by single particle electron cryo-microscopy | | Descriptor: | Transient receptor potential cation channel subfamily V member 1 | | Authors: | Wang, R.Y.-R, Barad, B.A, Fraser, J.S, DiMaio, F. | | Deposit date: | 2015-02-02 | | Release date: | 2015-09-02 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (3.275 Å) | | Cite: | EMRinger: side chain-directed model and map validation for 3D cryo-electron microscopy.
Nat.Methods, 12, 2015
|
|
6LCF
 
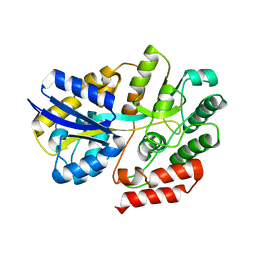 | | Crystal Structure of beta-L-arabinobiose binding protein - native | | Descriptor: | ABC transporter substrate binding component, beta-L-arabinofuranose-(1-2)-beta-L-arabinofuranose | | Authors: | Miyake, M, Arakawa, T, Fushinobu, S. | | Deposit date: | 2019-11-18 | | Release date: | 2020-04-22 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Structural analysis of beta-L-arabinobiose-binding protein in the metabolic pathway of hydroxyproline-rich glycoproteins in Bifidobacterium longum.
Febs J., 287, 2020
|
|
4OYE
 
 | | Crystal structure of GltPh R397A in apo | | Descriptor: | 425aa long hypothetical proton glutamate symport protein | | Authors: | Boudker, O, Oh, S. | | Deposit date: | 2014-02-11 | | Release date: | 2014-09-17 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (4 Å) | | Cite: | Coupled ion binding and structural transitions along the transport cycle of glutamate transporters.
Elife, 3, 2014
|
|
4OYF
 
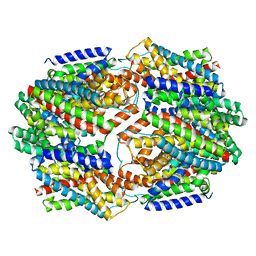 | | Crystal structure of GLTPH R397A IN Sodium-bound state | | Descriptor: | GLUTAMATE SYMPORT PROTEIN, SODIUM ION | | Authors: | Boudker, O, Oh, S, Verdon, G, Serio, R. | | Deposit date: | 2014-02-11 | | Release date: | 2014-08-13 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (3.41 Å) | | Cite: | Coupled ion binding and structural transitions along the transport cycle of glutamate transporters.
Elife, 3, 2014
|
|
3J4R
 
 | |
4OIR
 
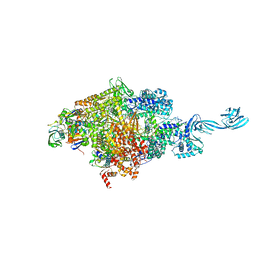 | | Crystal structure of Thermus thermophilus RNA polymerase transcription initiation complex soaked with GE23077 and rifamycin SV | | Descriptor: | (2Z)-2-methylbut-2-enoic acid, 5'-D(*CP*CP*T*GP*CP*AP*TP*CP*CP*GP*TP*GP*AP*GP*TP*CP*GP*AP*GP*GP*G)-3', 5'-D(*TP*AP*TP*AP*AP*TP*GP*GP*GP*AP*GP*CP*TP*GP*TP*CP*AP*CP*GP*GP*AP*TP*GP*CP*AP*GP*G)-3', ... | | Authors: | Zhang, Y, Ebright, R.H, Arnold, E. | | Deposit date: | 2014-01-20 | | Release date: | 2014-05-07 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.105 Å) | | Cite: | GE23077 binds to the RNA polymerase 'i' and 'i+1' sites and prevents the binding of initiating nucleotides.
Elife, 3, 2014
|
|
4O9L
 
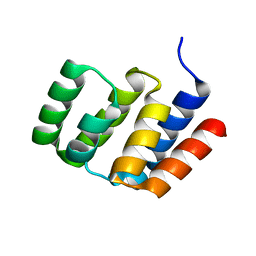 | |
6L0L
 
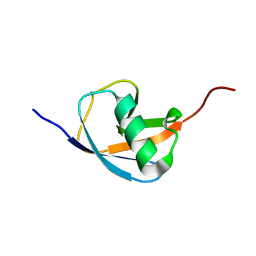 | | Hydra-1ubq de nova designed by Hydra based on ubiquitin | | Descriptor: | Hydra-1ubq | | Authors: | Ouyang, B. | | Deposit date: | 2019-09-26 | | Release date: | 2020-09-30 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Multiobjective heuristic algorithm for de novo protein design in a quantified continuous sequence space.
Comput Struct Biotechnol J, 19, 2021
|
|
4OIQ
 
 | | Crystal structure of Thermus thermophilus transcription initiation complex soaked with GE23077 and rifampicin | | Descriptor: | (2Z)-2-methylbut-2-enoic acid, 5'-D(*CP*CP*T*GP*CP*AP*TP*CP*CP*GP*TP*GP*AP*GP*TP*CP*GP*AP*GP*GP*G)-3', 5'-D(*TP*AP*TP*AP*AP*TP*GP*GP*GP*AP*GP*CP*TP*GP*TP*CP*AP*CP*GP*GP*AP*TP*GP*CP*AP*GP*G)-3', ... | | Authors: | Zhang, Y, Ebright, R.H, Arnold, E. | | Deposit date: | 2014-01-20 | | Release date: | 2014-05-07 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.624 Å) | | Cite: | GE23077 binds to the RNA polymerase 'i' and 'i+1' sites and prevents the binding of initiating nucleotides.
Elife, 3, 2014
|
|
4OLT
 
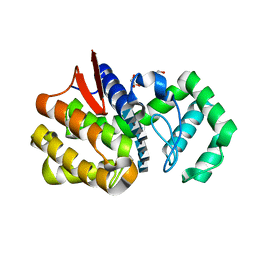 | | Chitosanase complex structure | | Descriptor: | 2-amino-2-deoxy-beta-D-glucopyranose-(1-4)-2-amino-2-deoxy-beta-D-glucopyranose-(1-4)-2-amino-2-deoxy-beta-D-glucopyranose-(1-4)-2-amino-2-deoxy-beta-D-glucopyranose-(1-4)-2-amino-2-deoxy-beta-D-glucopyranose-(1-4)-2-amino-2-deoxy-beta-D-glucopyranose, Chitosanase, GLYCEROL | | Authors: | Liu, W.Z, Lyu, Q.Q, Han, B.Q. | | Deposit date: | 2014-01-25 | | Release date: | 2014-04-30 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Structural insights into the substrate-binding mechanism for a novel chitosanase.
Biochem.J., 461, 2014
|
|
3J2Y
 
 | |
3J2W
 
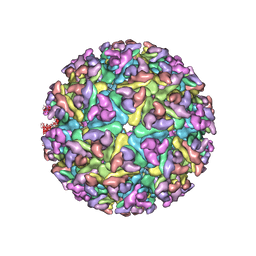 | | Electron cryo-microscopy of Chikungunya virus | | Descriptor: | Capsid protein, Glycoprotein E1, Glycoprotein E2 | | Authors: | Sun, S, Xiang, Y, Rossmann, M.G. | | Deposit date: | 2013-01-28 | | Release date: | 2013-04-24 | | Last modified: | 2018-07-18 | | Method: | ELECTRON MICROSCOPY (5 Å) | | Cite: | Structural analyses at pseudo atomic resolution of Chikungunya virus and antibodies show mechanisms of neutralization.
Elife, 2, 2013
|
|
4OIN
 
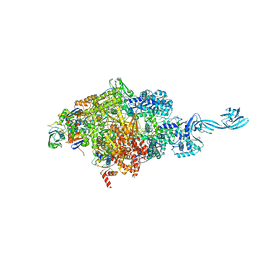 | | Crystal structure of Thermus thermophilus transcription initiation complex soaked with GE23077 | | Descriptor: | (2Z)-2-methylbut-2-enoic acid, 5'-D(*CP*CP*T*GP*CP*AP*TP*CP*CP*GP*TP*GP*AP*GP*TP*CP*GP*AP*G)-3', 5'-D(*TP*AP*TP*AP*AP*TP*GP*GP*GP*AP*GP*CP*TP*GP*TP*CP*AP*CP*GP*GP*AP*TP*GP*CP*AP*GP*G)-3', ... | | Authors: | Zhang, Y, Ebright, R.H, Arnold, E. | | Deposit date: | 2014-01-20 | | Release date: | 2014-05-07 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | GE23077 binds to the RNA polymerase 'i' and 'i+1' sites and prevents the binding of initiating nucleotides.
Elife, 3, 2014
|
|
4O9F
 
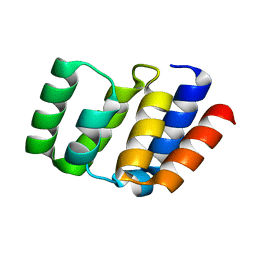 | |
4OIO
 
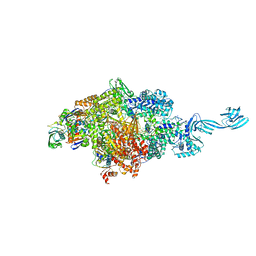 | | Crystal structure of Thermus thermophilus pre-insertion substrate complex for de novo transcription initiation | | Descriptor: | 5'-D(*CP*CP*TP*GP*CP*AP*TP*CP*CP*GP*TP*GP*AP*GP*TP*CP*GP*AP*GP*GP*G)-3', 5'-D(*TP*AP*TP*AP*AP*TP*GP*GP*GP*AP*GP*CP*TP*GP*TP*CP*AP*CP*GP*GP*AP*TP*GP*CP*AP*GP*G)-3', 5'-O-[(S)-hydroxy{[(S)-hydroxy(phosphonooxy)phosphoryl]methyl}phosphoryl]cytidine, ... | | Authors: | Zhang, Y, Ebright, R.H, Arnold, E. | | Deposit date: | 2014-01-20 | | Release date: | 2014-05-07 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | GE23077 binds to the RNA polymerase 'i' and 'i+1' sites and prevents the binding of initiating nucleotides.
Elife, 3, 2014
|
|
4P1A
 
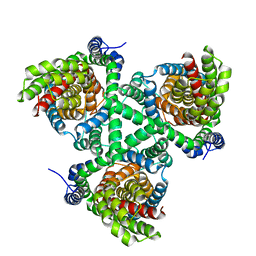 | |
4P19
 
 | |
4P3J
 
 | |
4P0T
 
 | |
4P6H
 
 | |
6MKM
 
 | | Crystallographic solvent mapping analysis of DMSO/Tris bound to APE1 | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, DIMETHYL SULFOXIDE, ... | | Authors: | Georgiadis, M.M, He, H, Chen, Q. | | Deposit date: | 2018-09-25 | | Release date: | 2019-01-30 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.673 Å) | | Cite: | Discovery of Macrocyclic Inhibitors of Apurinic/Apyrimidinic Endonuclease 1.
J. Med. Chem., 62, 2019
|
|
4POF
 
 | | PfMCM N-terminal domain without DNA | | Descriptor: | Cell division control protein 21, ZINC ION | | Authors: | Froelich, C.A, Kang, S, Epling, L.B, Bell, S.P, Enemark, E.J. | | Deposit date: | 2014-02-25 | | Release date: | 2014-04-09 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.648 Å) | | Cite: | A conserved MCM single-stranded DNA binding element is essential for replication initiation.
Elife, 3, 2014
|
|
4P6Z
 
 | |
6MK3
 
 | | Crystallographic solvent mapping analysis of DMSO bound to APE1 | | Descriptor: | 1,2-ETHANEDIOL, DIMETHYL SULFOXIDE, DNA-(apurinic or apyrimidinic site) lyase | | Authors: | Georgiadis, M.M, He, H, Chen, Q. | | Deposit date: | 2018-09-24 | | Release date: | 2019-01-30 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.478 Å) | | Cite: | Discovery of Macrocyclic Inhibitors of Apurinic/Apyrimidinic Endonuclease 1.
J. Med. Chem., 62, 2019
|
|
6MKO
 
 | |
