6L76
 
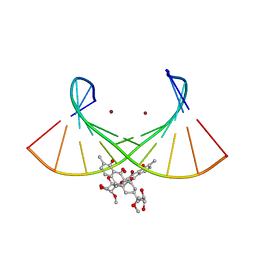 | | Crystal structure of the Ni(II)(Chro)2-d(TTGGGCCGAA/TTCGGCCCAA) complex at 2.94 angstrom resolution | | Descriptor: | (1S)-5-deoxy-1-O-methyl-1-C-[(2R,3S)-3,5,7,10-tetrahydroxy-6-methyl-4-oxo-1,2,3,4-tetrahydroanthracen-2-yl]-D-xylulose, 2,6-dideoxy-4-O-methyl-alpha-D-galactopyranose-(1-3)-(2R,3R,6R)-6-hydroxy-2-methyltetrahydro-2H-pyran-3-yl acetate, 3-C-methyl-4-O-acetyl-alpha-L-Olivopyranose-(1-3)-(2R,5S,6R)-6-methyltetrahydro-2H-pyran-2,5-diol-(1-3)-(2R,5S,6R)-6-methyltetrahydro-2H-pyran-2,5-diol, ... | | Authors: | Hou, M.H, Jhan, C.R, Satange, R.B, Lin, S.M. | | Deposit date: | 2019-10-31 | | Release date: | 2021-01-13 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.94 Å) | | Cite: | Targeting the ALS/FTD-associated A-DNA kink with anthracene-based metal complex causes DNA backbone straightening and groove contraction.
Nucleic Acids Res., 49, 2021
|
|
6J0I
 
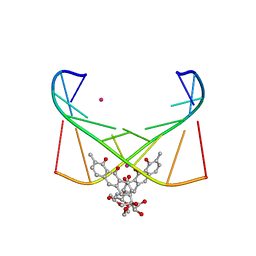 | | Structure of [Co2+-(Chromomycin A3)2]-d(TTGGCGAA)2 complex | | Descriptor: | 1,2-HYDRO-1-OXY-3,4-HYDRO-3-(1-METHOXY-2-OXY-3,4-DIHYDROXYPENTYL)-8,9-DIHYROXY-7-METHYLANTHRACENE, 2,6-dideoxy-4-O-methyl-alpha-D-galactopyranose-(1-3)-(2R,3R,6R)-6-hydroxy-2-methyltetrahydro-2H-pyran-3-yl acetate, 3-C-methyl-4-O-acetyl-alpha-L-Olivopyranose-(1-3)-(2R,5S,6R)-6-methyltetrahydro-2H-pyran-2,5-diol-(1-3)-(2R,5S,6R)-6-methyltetrahydro-2H-pyran-2,5-diol, ... | | Authors: | Satange, R.B, Chuang, C.Y, Hou, M.H. | | Deposit date: | 2018-12-24 | | Release date: | 2019-07-24 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Polymorphic G:G mismatches act as hotspots for inducing right-handed Z DNA by DNA intercalation.
Nucleic Acids Res., 47, 2019
|
|
6HN0
 
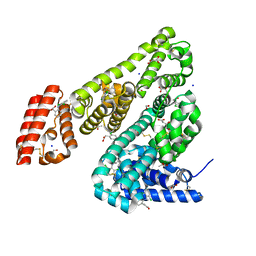 | | Complex of Ovine Serum Albumin with diclofenac | | Descriptor: | (2S)-2-hydroxybutanedioic acid, 2-[2,6-DICHLOROPHENYL)AMINO]BENZENEACETIC ACID, ACETATE ION, ... | | Authors: | Talaj, J.A, Bujacz, A, Bujacz, G. | | Deposit date: | 2018-09-13 | | Release date: | 2019-10-09 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | Structural Investigation of Diclofenac Binding to Ovine, Caprine, and Leporine Serum Albumins.
Int J Mol Sci, 24, 2023
|
|
6H9B
 
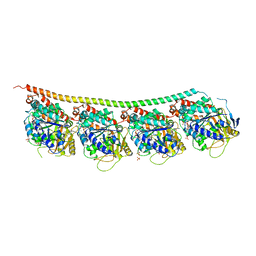 | | 1,1-Diheterocyclic Ethylenes Derived from Quinaldine and Carbazole as New Tubulin Polymerization Inhibitors: Synthesis, Metabolism, and Biological Evaluation | | Descriptor: | 9-methyl-3-[1-(2-methylquinolin-4-yl)ethenyl]carbazole, GUANOSINE-5'-DIPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Varela, P.F, Gigant, B. | | Deposit date: | 2018-08-03 | | Release date: | 2018-12-19 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | 1,1-Diheterocyclic Ethylenes Derived from Quinaldine and Carbazole as New Tubulin-Polymerization Inhibitors: Synthesis, Metabolism, and Biological Evaluation.
J. Med. Chem., 62, 2019
|
|
6GX7
 
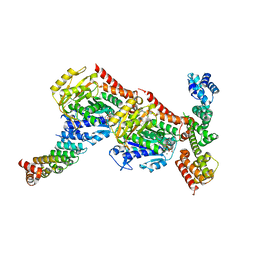 | | Tubulin-CopN-alphaRep complex | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, GLYCEROL, GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Gigant, B, Campanacci, V. | | Deposit date: | 2018-06-26 | | Release date: | 2019-04-24 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.19 Å) | | Cite: | Insight into microtubule nucleation from tubulin-capping proteins.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
6GWD
 
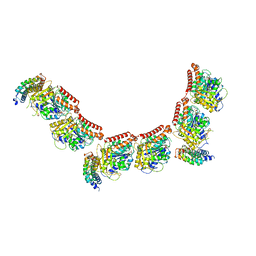 | | Tubulin:iiH5 alphaRep complex | | Descriptor: | ALPHA-TUBULIN, GUANOSINE-5'-DIPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Gigant, B, Campanacci, V. | | Deposit date: | 2018-06-22 | | Release date: | 2019-01-30 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Selection and Characterization of Artificial Proteins Targeting the Tubulin alpha Subunit.
Structure, 27, 2019
|
|
6GWC
 
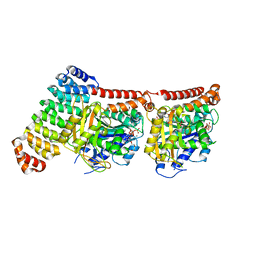 | | Tubulin:iE5 alphaRep complex | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, ALPHA-TUBULIN, GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Gigant, B, Campanacci, V. | | Deposit date: | 2018-06-22 | | Release date: | 2019-01-30 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Selection and Characterization of Artificial Proteins Targeting the Tubulin alpha Subunit.
Structure, 27, 2019
|
|
6GVN
 
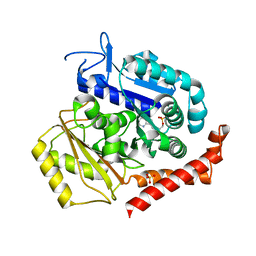 | | Tubulin:TM-3 DARPin complex | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, GLYCEROL, GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Gigant, B, Cantos Fernandes, S, Campanacci, V. | | Deposit date: | 2018-06-21 | | Release date: | 2019-04-24 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.69 Å) | | Cite: | Insight into microtubule nucleation from tubulin-capping proteins.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
6GVM
 
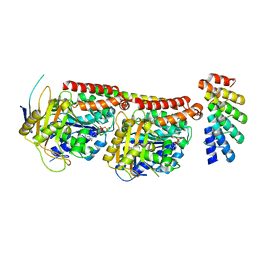 | | Tubulin:F3II DARPin complex | | Descriptor: | F3II DARPIN, GUANOSINE-5'-DIPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Gigant, B, Campanacci, V, Cantos Fernandes, S. | | Deposit date: | 2018-06-21 | | Release date: | 2019-04-24 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Insight into microtubule nucleation from tubulin-capping proteins.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
6CEB
 
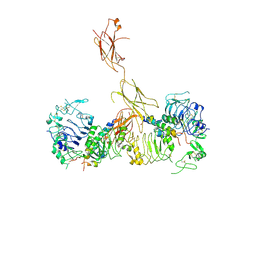 | | Insulin Receptor ectodomain in complex with two insulin molecules - C1 symmetry | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Scapin, G, Dandey, V.P, Zhang, Z, Strickland, C, Potter, C.S, Carragher, B. | | Deposit date: | 2018-02-11 | | Release date: | 2018-03-14 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (4.7 Å) | | Cite: | Structure of the insulin receptor-insulin complex by single-particle cryo-EM analysis.
Nature, 556, 2018
|
|
6CE9
 
 | | Insulin Receptor ectodomain in complex with two insulin molecules | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Scapin, G, Dandey, V.P, Zhang, Z, Strickland, C, Potter, C.S, Carragher, B. | | Deposit date: | 2018-02-11 | | Release date: | 2018-03-14 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Structure of the insulin receptor-insulin complex by single-particle cryo-EM analysis.
Nature, 556, 2018
|
|
6CE7
 
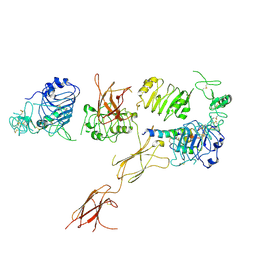 | | Insulin Receptor ectodomain in complex with one insulin molecule | | Descriptor: | Insulin A chain, Insulin B chain, Insulin receptor, ... | | Authors: | Scapin, G, Dandey, V.P, Zhang, Z, Strickland, C, Potter, C.S, Carragher, B. | | Deposit date: | 2018-02-11 | | Release date: | 2018-03-14 | | Last modified: | 2019-12-18 | | Method: | ELECTRON MICROSCOPY (7.4 Å) | | Cite: | Structure of the insulin receptor-insulin complex by single-particle cryo-EM analysis.
Nature, 556, 2018
|
|
5Z4V
 
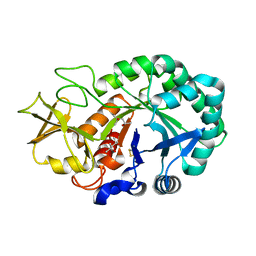 | | Crystal structure of the sheep signalling glycoprotein (SPS-40) complex with 2-methyl-2-4-pentanediol at 1.65A resolution reveals specific binding characteristics of SPS-40 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, Chitinase-3-like protein 1 | | Authors: | Sharma, P, Singh, P.K, Singh, N, Sharma, S, Kaur, P, Betzel, C, Singh, T.P. | | Deposit date: | 2018-01-15 | | Release date: | 2018-02-14 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Crystal structure of the sheep signalling glycoprotein (SPS-40) complex with 2-methyl-2-4-pentanediol at 1.65A resolution reveals specific binding characteristics of SPS-40
To Be Published
|
|
5Z2O
 
 | |
5Z26
 
 | |
5YZE
 
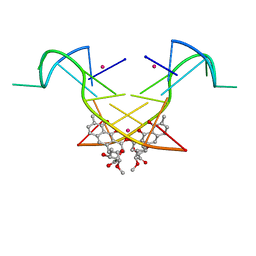 | | Crystal structure of the [Co2+-(chromomycin A3)2]-d(CCG)3 complex | | Descriptor: | (1S)-5-deoxy-1-O-methyl-1-C-[(2R,3S)-3,5,7,10-tetrahydroxy-6-methyl-4-oxo-1,2,3,4-tetrahydroanthracen-2-yl]-D-xylulose, 2,6-dideoxy-4-O-methyl-alpha-D-galactopyranose-(1-3)-(2R,3R,6R)-6-hydroxy-2-methyltetrahydro-2H-pyran-3-yl acetate, 3-C-methyl-4-O-acetyl-alpha-L-Olivopyranose-(1-3)-(2R,5S,6R)-6-methyltetrahydro-2H-pyran-2,5-diol-(1-3)-(2R,5S,6R)-6-methyltetrahydro-2H-pyran-2,5-diol, ... | | Authors: | Hou, M.H, Chen, Y.W, Wu, P.C, Satange, R.B. | | Deposit date: | 2017-12-14 | | Release date: | 2018-10-17 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | CoII(Chromomycin)2 Complex Induces a Conformational Change of CCG Repeats from i-Motif to Base-Extruded DNA Duplex
Int J Mol Sci, 19, 2018
|
|
5XEW
 
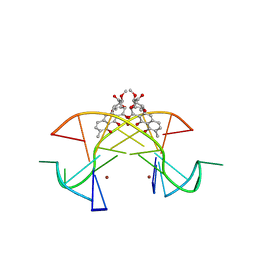 | | Crystal structure of the [Ni2+-(chromomycin A3)2]-CCG repeats complex | | Descriptor: | (1S)-5-deoxy-1-O-methyl-1-C-[(2R,3S)-3,5,7,10-tetrahydroxy-6-methyl-4-oxo-1,2,3,4-tetrahydroanthracen-2-yl]-D-xylulose, 2,6-dideoxy-4-O-methyl-alpha-D-galactopyranose-(1-3)-(2R,3R,6R)-6-hydroxy-2-methyltetrahydro-2H-pyran-3-yl acetate, 3-C-methyl-4-O-acetyl-alpha-L-Olivopyranose-(1-3)-(2R,5S,6R)-6-methyltetrahydro-2H-pyran-2,5-diol-(1-3)-(2R,5S,6R)-6-methyltetrahydro-2H-pyran-2,5-diol, ... | | Authors: | Tseng, W.H, Wu, P.C, Hou, M.H. | | Deposit date: | 2017-04-06 | | Release date: | 2017-06-21 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.751 Å) | | Cite: | Induced-Fit Recognition of CCG Trinucleotide Repeats by a Nickel-Chromomycin Complex Resulting in Large-Scale DNA Deformation
Angew. Chem. Int. Ed. Engl., 56, 2017
|
|
5WBE
 
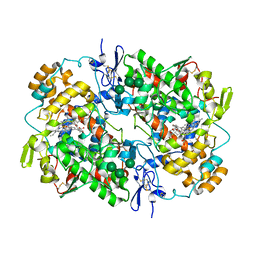 | | COX-1:MOFEZOLAC COMPLEX STRUCTURE | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Mofezolac, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Cingolani, G, Panella, A, Perrone, M.G, Vitale, P, Smith, W.L, Scilimati, A. | | Deposit date: | 2017-06-28 | | Release date: | 2017-07-26 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Structural basis for selective inhibition of Cyclooxygenase-1 (COX-1) by diarylisoxazoles mofezolac and 3-(5-chlorofuran-2-yl)-5-methyl-4-phenylisoxazole (P6).
Eur J Med Chem, 138, 2017
|
|
5UDH
 
 | | HHARI/ARIH1-UBCH7~Ubiquitin | | Descriptor: | E3 ubiquitin-protein ligase ARIH1, Ubiquitin C variant, Ubiquitin-conjugating enzyme E2 L3, ... | | Authors: | Miller, D.J, Schulman, B.A. | | Deposit date: | 2016-12-27 | | Release date: | 2017-06-14 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (3.24 Å) | | Cite: | Structural Studies of HHARI/UbcH7Ub Reveal Unique E2Ub Conformational Restriction by RBR RING1.
Structure, 25, 2017
|
|
5U6X
 
 | | COX-1:P6 COMPLEX STRUCTURE | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 3-(5-chlorofuran-2-yl)-5-methyl-4-phenyl-1,2-oxazole, PROTOPORPHYRIN IX CONTAINING FE, ... | | Authors: | Cingolani, G, Panella, A, Perrone, M.G, Vitale, P, Smith, W.L, Scilimati, A. | | Deposit date: | 2016-12-09 | | Release date: | 2017-08-09 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.93 Å) | | Cite: | Structural basis for selective inhibition of Cyclooxygenase-1 (COX-1) by diarylisoxazoles mofezolac and 3-(5-chlorofuran-2-yl)-5-methyl-4-phenylisoxazole (P6).
Eur J Med Chem, 138, 2017
|
|
5TTE
 
 | | Crystal Structure of an RBR E3 ubiquitin ligase in complex with an E2-Ub thioester intermediate mimic | | Descriptor: | E3 ubiquitin-protein ligase ARIH1, Ubiquitin-conjugating enzyme E2 L3, ZINC ION, ... | | Authors: | Yuan, L, Lv, Z, Olsen, S.K. | | Deposit date: | 2016-11-03 | | Release date: | 2017-08-23 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.501 Å) | | Cite: | Structural insights into the mechanism and E2 specificity of the RBR E3 ubiquitin ligase HHARI.
Nat Commun, 8, 2017
|
|
5OUK
 
 | | Crystal structure of human AKR1B1 complexed with NADP+ and compound 41 | | Descriptor: | 2-[9-fluoranyl-5-(4-methoxyphenyl)-3-methyl-1-oxidanylidene-pyrimido[4,5-c]quinolin-2-yl]ethanoic acid, Aldose reductase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Cousido-Siah, A, Ruiz, F.X, Mitschler, A, Metwally, K, Podjarny, A. | | Deposit date: | 2017-08-24 | | Release date: | 2018-05-09 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (0.959 Å) | | Cite: | Design, synthesis, structure-activity relationships and X-ray structural studies of novel 1-oxopyrimido[4,5-c]quinoline-2-acetic acid derivatives as selective and potent inhibitors of human aldose reductase.
Eur J Med Chem, 152, 2018
|
|
5OUJ
 
 | | Crystal structure of human AKR1B1 complexed with NADP+ and compound 39 | | Descriptor: | 2-[(1~{R})-5-(4-chlorophenyl)-9-fluoranyl-3-methyl-1-oxidanyl-1~{H}-pyrimido[4,5-c]quinolin-2-yl]ethanoic acid, Aldose reductase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Cousido-Siah, A, Ruiz, F.X, Mitschler, A, Metwally, K, Podjarny, A. | | Deposit date: | 2017-08-24 | | Release date: | 2018-05-09 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (0.96 Å) | | Cite: | Design, synthesis, structure-activity relationships and X-ray structural studies of novel 1-oxopyrimido[4,5-c]quinoline-2-acetic acid derivatives as selective and potent inhibitors of human aldose reductase.
Eur J Med Chem, 152, 2018
|
|
5OU0
 
 | | Crystal structure of human AKR1B1 complexed with NADP+ and compound 37 | | Descriptor: | 2-[5-(4-chlorophenyl)-3-methyl-1-oxidanylidene-pyrimido[4,5-c]quinolin-2-yl]ethanoic acid, Aldose reductase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Cousido-Siah, A, Ruiz, F.X, Mitschler, A, Metwally, K, Podjarny, A. | | Deposit date: | 2017-08-23 | | Release date: | 2018-05-09 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (0.94 Å) | | Cite: | Design, synthesis, structure-activity relationships and X-ray structural studies of novel 1-oxopyrimido[4,5-c]quinoline-2-acetic acid derivatives as selective and potent inhibitors of human aldose reductase.
Eur J Med Chem, 152, 2018
|
|
5ORF
 
 | | Structure of ovine serum albumin in P1 space group | | Descriptor: | DI(HYDROXYETHYL)ETHER, PROLINE, Serum albumin, ... | | Authors: | Talaj, J.A, Bujacz, A, Bujacz, G, Pietrzyk-Brzezinska, A.J. | | Deposit date: | 2017-08-16 | | Release date: | 2017-11-15 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.54 Å) | | Cite: | Crystal structures of serum albumins from domesticated ruminants and their complexes with 3,5-diiodosalicylic acid.
Acta Crystallogr D Struct Biol, 73, 2017
|
|
