2MV8
 
 | | Solution structure of Ovis Aries PrP with mutation delta190-197 | | Descriptor: | Major prion protein | | Authors: | Munoz, C, Egalon, A, Beringue, V, Rezaei, H, Dron, M, Sizun, C. | | Deposit date: | 2014-09-25 | | Release date: | 2015-10-07 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Generating Bona Fide Mammalian Prions with Internal Deletions.
J.Virol., 90, 2016
|
|
2M9Y
 
 | |
2M1J
 
 | | Ovine Doppel Signal peptide (1-30) | | Descriptor: | Prion-like protein doppel | | Authors: | Pimenta, J, Viegas, A, Sardinha, J, Santos, A, Cantante, C, Dias, F.M.V, Soares, R, Cabrita, E.J, Fontes, C.M.G.A, Prates, J.A.M, Pereira, R.M.L.N. | | Deposit date: | 2012-11-28 | | Release date: | 2013-10-16 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | NMR solution structure and SRP54M predicted interaction of the N-terminal sequence (1-30) of the ovine Doppel protein.
Peptides, 49C, 2013
|
|
2KTM
 
 | | Solution NMR structure of H2H3 domain of ovine prion protein (residues 167-234) | | Descriptor: | Major prion protein | | Authors: | Pastore, A, Adrover, M, Pauwels, K, de Chiara, C, Prigent, S, Rezeai, H. | | Deposit date: | 2010-02-04 | | Release date: | 2010-04-07 | | Last modified: | 2021-11-10 | | Method: | SOLUTION NMR | | Cite: | Prion fibrillization is mediated by a native structural element that comprises helices H2 and H3.
J.Biol.Chem., 285, 2010
|
|
2ISF
 
 | |
2IS7
 
 | | Crystal Structure of Aldose Reductase complexed with Dichlorophenylacetic acid | | Descriptor: | (2,6-DICHLOROPHENYL)ACETIC ACID, Aldose reductase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Harrison, D.H.T, Milne, A, Brownlee, J.M. | | Deposit date: | 2006-10-16 | | Release date: | 2006-11-21 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural and thermodynamic studies of simple aldose reductase-inhibitor complexes.
Bioorg.Chem., 34, 2006
|
|
2IQD
 
 | | Crystal Structure of Aldose Reductase complexed with Lipoic Acid | | Descriptor: | Aldose reductase, LIPOIC ACID, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Harrison, D.H.T, Carlson, E, Pape, E, Brownlee, J.M. | | Deposit date: | 2006-10-13 | | Release date: | 2006-11-28 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural and thermodynamic studies of simple aldose reductase-inhibitor complexes.
Bioorg.Chem., 34, 2006
|
|
2IQ0
 
 | |
2IPW
 
 | | Crystal Structure of C298A W219Y Aldose Reductase complexed with Dichlorophenylacetic acid | | Descriptor: | (2,6-DICHLOROPHENYL)ACETIC ACID, Aldose reductase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Harrison, D.H.T, Pape, E, Brownlee, J.M. | | Deposit date: | 2006-10-12 | | Release date: | 2006-11-28 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural and thermodynamic studies of simple aldose reductase-inhibitor complexes.
Bioorg.Chem., 34, 2006
|
|
2INZ
 
 | | Crystal Structure of Aldose Reductase complexed with 2-Hydroxyphenylacetic Acid | | Descriptor: | (2-HYDROXYPHENYL)ACETIC ACID, Aldose reductase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Harrison, D.H.T, Pape, E, Carlson, E, Brownlee, J.M. | | Deposit date: | 2006-10-09 | | Release date: | 2006-11-21 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural and thermodynamic studies of simple aldose reductase-inhibitor complexes.
Bioorg.Chem., 34, 2006
|
|
2INE
 
 | |
2IKJ
 
 | | Human aldose reductase complexed with nitro-substituted IDD-type inhibitor | | Descriptor: | (5-CHLORO-2-{[(3-NITROBENZYL)AMINO]CARBONYL}PHENOXY)ACETIC ACID, Aldose reductase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Steuber, H, Koch, C, Heine, A, Klebe, G. | | Deposit date: | 2006-10-02 | | Release date: | 2007-05-01 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structural and Thermodynamic Study on Aldose Reductase: Nitro-substituted Inhibitors with Strong Enthalpic Binding Contribution
J.Mol.Biol., 368, 2007
|
|
2IKI
 
 | | Human aldose reductase complexed with halogenated IDD-type inhibitor | | Descriptor: | (2-{[(4-BROMO-2-FLUOROBENZYL)AMINO]CARBONYL}-5-CHLOROPHENOXY)ACETIC ACID, Aldose reductase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Steuber, H, Koch, C, Heine, A, Klebe, G. | | Deposit date: | 2006-10-02 | | Release date: | 2007-05-01 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | Structural and Thermodynamic Study on Aldose Reductase: Nitro-substituted Inhibitors with Strong Enthalpic Binding Contribution
J.Mol.Biol., 368, 2007
|
|
2IKH
 
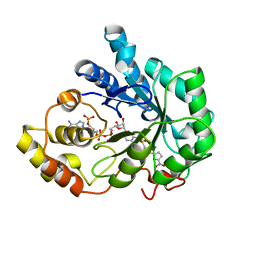 | | Human aldose reductase complexed with nitrofuryl-oxadiazol inhibitor at 1.55 A | | Descriptor: | Aldose reductase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, {[5-(5-NITRO-2-FURYL)-1,3,4-OXADIAZOL-2-YL]THIO}ACETIC ACID | | Authors: | Steuber, H, Koch, C, Heine, A, Klebe, G. | | Deposit date: | 2006-10-02 | | Release date: | 2007-05-01 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structural and Thermodynamic Study on Aldose Reductase: Nitro-substituted Inhibitors with Strong Enthalpic Binding Contribution
J.Mol.Biol., 368, 2007
|
|
2IKG
 
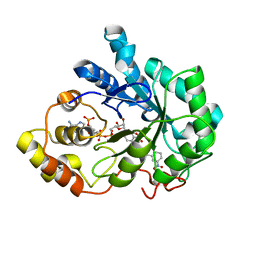 | | Aldose reductase complexed with nitrophenyl-oxadiazol type inhibitor at 1.43 A | | Descriptor: | 4-[3-(3-NITROPHENYL)-1,2,4-OXADIAZOL-5-YL]BUTANOIC ACID, Aldose reductase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Steuber, H, Koch, C, Heine, A, Klebe, G. | | Deposit date: | 2006-10-02 | | Release date: | 2007-05-01 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.43 Å) | | Cite: | Structural and Thermodynamic Study on Aldose Reductase: Nitro-substituted Inhibitors with Strong Enthalpic Binding Contribution
J.Mol.Biol., 368, 2007
|
|
2IKC
 
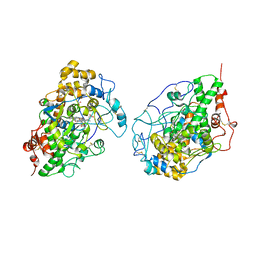 | | Crystal structure of sheep lactoperoxidase at 3.25 A resolution reveals the binding sites for formate | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Sheikh, I.A, Singh, N, Singh, A.K, Sharma, S, Singh, T.P. | | Deposit date: | 2006-10-02 | | Release date: | 2006-10-17 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (3.25 Å) | | Cite: | Crystal structure of sheep lactoperoxidase at 3.25 A resolution reveals the binding sites for formate
To be Published
|
|
2HLR
 
 | |
2HLQ
 
 | |
2H1W
 
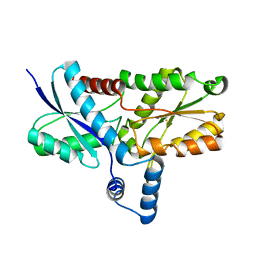 | | Crystal structure of the His183Ala mutant variant of Bacillus subtilis ferrochelatase | | Descriptor: | FE (II) ION, Ferrochelatase, MAGNESIUM ION | | Authors: | Hansson, M.D, Karlberg, T, Arys Rahardja, M, Al-Karadaghi, S, Hansson, M. | | Deposit date: | 2006-05-17 | | Release date: | 2007-01-16 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Amino Acid Residues His183 and Glu264 in Bacillus subtilis Ferrochelatase Direct and Facilitate the Insertion of Metal Ion into Protoporphyrin IX
Biochemistry, 46, 2007
|
|
2H1V
 
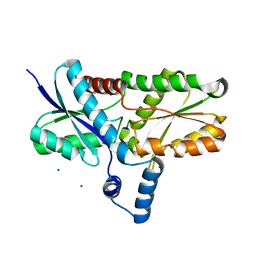 | | Crystal structure of the Lys87Ala mutant variant of Bacillus subtilis ferrochelatase | | Descriptor: | Ferrochelatase, MAGNESIUM ION | | Authors: | Hansson, M.D, Karlberg, T, Arys Rahardja, M, Al-Karadaghi, S, Hansson, M. | | Deposit date: | 2006-05-17 | | Release date: | 2007-01-16 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Amino Acid Residues His183 and Glu264 in Bacillus subtilis Ferrochelatase Direct and Facilitate the Insertion of Metal Ion into Protoporphyrin IX
Biochemistry, 46, 2007
|
|
2G8Z
 
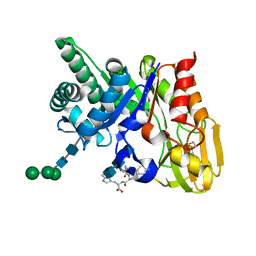 | | Crystal structure of the ternary complex of signalling protein from sheep (SPS-40) with trimer and designed peptide at 2.5A resolution | | Descriptor: | (TRP)(PRO)(TRP), 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Chitinase-3-like protein 1, ... | | Authors: | Ethayathulla, A.S, Srivastava, D.B, Kumar, J, Somvanshi, R.K, Sharma, S, Singh, T.P. | | Deposit date: | 2006-03-04 | | Release date: | 2006-04-04 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of the ternary complex of signalling protein from sheep (SPS-40) with trimer and designed peptide at 2.5A resolution
To be Published
|
|
2G41
 
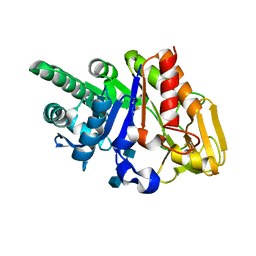 | | Crystal structure of the complex of sheep signalling glycoprotein with chitin trimer at 3.0A resolution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, SIGNAL PROCESSING PROTEIN, beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Srivastava, D.B, Ethayathulla, A.S, Kumar, J, Bhushan, A, Sharma, S, Singh, T.P. | | Deposit date: | 2006-02-21 | | Release date: | 2006-04-04 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Carbohydrate binding properties and carbohydrate induced conformational switch in sheep secretory glycoprotein (SPS-40): crystal structures of four complexes of SPS-40 with chitin-like oligosaccharides
J.Struct.Biol., 158, 2007
|
|
2FDM
 
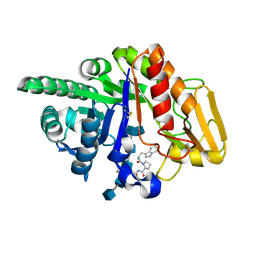 | | Crystal structure of the ternary complex of signalling glycoprotein frm sheep (SPS-40)with hexasaccharide (NAG6) and peptide Trp-Pro-Trp at 3.0A resolution | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-alpha-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-alpha-D-glucopyranose, Chitinase-3-like protein 1, ... | | Authors: | Srivastava, D.B, Ethayathulla, A.S, Kumar, J, Somvanshi, R.K, Sharma, S, Singh, T.P. | | Deposit date: | 2005-12-14 | | Release date: | 2006-01-10 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal structure of the ternary complex of signalling glycoprotein frm sheep (SPS-40)
with hexasaccharide (NAG6) and peptide Trp-Pro-Trp at 3.0A resolution
To be Published
|
|
2F8K
 
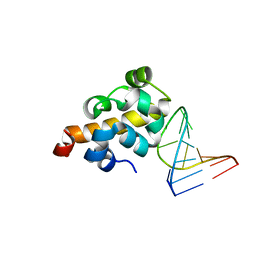 | | Sequence specific recognition of RNA hairpins by the SAM domain of Vts1 | | Descriptor: | 5'-R(*UP*AP*AP*UP*CP*UP*UP*UP*GP*AP*CP*AP*GP*AP*UP*U)-3', Protein VTS1 | | Authors: | Aviv, T, Lin, Z, Ben-Ari, G, Smibert, C.A, Sicheri, F. | | Deposit date: | 2005-12-02 | | Release date: | 2006-01-24 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Sequence-specific recognition of RNA hairpins by the SAM domain of Vts1p.
Nat.Struct.Mol.Biol., 13, 2006
|
|
2DSW
 
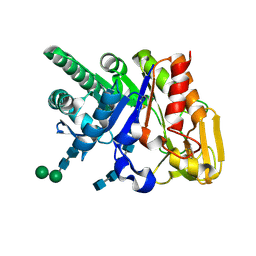 | | Binding of chitin-like polysaccharides to protective signalling factor: crystal structure of the complex of signalling protein from sheep (SPS-40) with a pentasaccharide at 2.8 A resolution | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Chitinase-3-like protein 1, alpha-D-mannopyranose-(1-4)-alpha-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Srivastava, D.B, Ethayathulla, A.S, Kumar, J, Singh, N, Sharma, S, Kaur, P, Bhushan, A, Singh, T.P. | | Deposit date: | 2006-07-07 | | Release date: | 2006-08-01 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Carbohydrate binding properties and carbohydrate induced conformational switch in sheep secretory glycoprotein (SPS-40): crystal structures of four complexes of SPS-40 with chitin-like oligosaccharides
J.Struct.Biol., 158, 2007
|
|
