6SAB
 
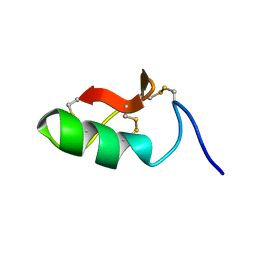 | | M-BUTX-Ptr1a (Parabuthus transvaalicus) | | Descriptor: | M-BUTX-Ptr1a | | Authors: | Meudal, H, Landon, C, Delmas, A.F. | | Deposit date: | 2019-07-16 | | Release date: | 2020-07-22 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | A Venomics Approach Coupled to High-Throughput Toxin Production Strategies Identifies the First Venom-Derived Melanocortin Receptor Agonists.
J.Med.Chem., 63, 2020
|
|
5B5J
 
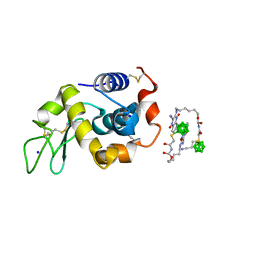 | | Hen egg white lysozyme with boron tracedrug UTX-97 | | Descriptor: | 2-cyano-3-((6-(((2-((2-cyanoethyl)(borocaptate-10B)sulfonio)acetyl)carbamoyl)oxy)hexyl)amino)quinoxaline 1,4-dioxide, Lysozyme C, SODIUM ION | | Authors: | Morimoto, Y. | | Deposit date: | 2016-05-11 | | Release date: | 2017-06-28 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.26 Å) | | Cite: | Structural Insight Into Protein Binding of Boron Tracedrug UTX-97 Revealed by the Co-Crystal Structure With Lysozyme at 1.26 angstrom Resolution.
J Pharm Sci, 105, 2016
|
|
3AVR
 
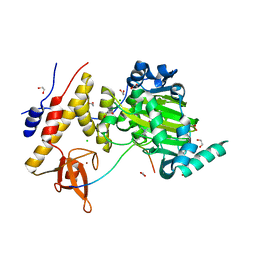 | | Catalytic fragment of UTX/KDM6A bound with histone H3K27me3 peptide, N-oxyalylglycine, and Ni(II) | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Histone H3, ... | | Authors: | Sengoku, T, Yokoyama, S. | | Deposit date: | 2011-03-07 | | Release date: | 2011-10-19 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.803 Å) | | Cite: | Structural basis for histone H3 Lys 27 demethylation by UTX/KDM6A
Genes Dev., 25, 2011
|
|
3AVS
 
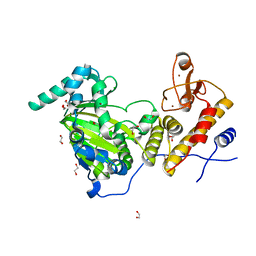 | | Catalytic fragment of UTX/KDM6A bound with N-oxyalylglycine, and Ni(II) | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Lysine-specific demethylase 6A, ... | | Authors: | Sengoku, T, Yokoyama, S. | | Deposit date: | 2011-03-07 | | Release date: | 2011-10-19 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural basis for histone H3 Lys 27 demethylation by UTX/KDM6A
Genes Dev., 25, 2011
|
|
5EXV
 
 | |
1WT7
 
 | | Solution structure of BuTX-MTX: a butantoxin-maurotoxin chimera | | Descriptor: | BuTX-MTX | | Authors: | M'Barek, S, Chagot, B, Andreotti, N, Visan, V, Mansuelle, P, Grissmer, S, Marrakchi, M, El Ayeb, M, Sampieri, F, Darbon, H, Fajloun, Z, De Waard, M, Sabatier, J.-M. | | Deposit date: | 2004-11-16 | | Release date: | 2004-11-30 | | Last modified: | 2024-10-30 | | Method: | SOLUTION NMR | | Cite: | Increasing the molecular contacts between maurotoxin and Kv1.2 channel augments ligand affinity.
Proteins, 60, 2005
|
|
6FUL
 
 | | Crystal structure of UTX complexed with 5-hydroxy-4-keto-1-methyl-picolinate | | Descriptor: | 1-methyl-5-oxidanyl-4-oxidanylidene-pyridine-2-carboxylic acid, 2-(2-METHOXYETHOXY)ETHANOL, Lysine-specific demethylase 6A, ... | | Authors: | Esposito, C, Sledz, P, Caflisch, A. | | Deposit date: | 2018-02-27 | | Release date: | 2018-10-10 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.649 Å) | | Cite: | In Silico Identification of JMJD3 Demethylase Inhibitors.
J Chem Inf Model, 58, 2018
|
|
6G8F
 
 | | Crystal structure of UTX complexed with GSK-J1 | | Descriptor: | 3-[[2-pyridin-2-yl-6-(1,2,4,5-tetrahydro-3-benzazepin-3-yl)pyrimidin-4-yl]amino]propanoic acid, CHLORIDE ION, DIMETHYL SULFOXIDE, ... | | Authors: | Esposito, C, Sledz, P, Caflisch, A. | | Deposit date: | 2018-04-08 | | Release date: | 2018-10-10 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.043 Å) | | Cite: | In Silico Identification of JMJD3 Demethylase Inhibitors.
J Chem Inf Model, 58, 2018
|
|
6FUK
 
 | | Crystal structure of UTX complexed with 5-carboxy-8-hydroxyquinoline | | Descriptor: | 2-(2-METHOXYETHOXY)ETHANOL, 8-hydroxyquinoline-5-carboxylic acid, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Esposito, C, Sledz, P, Caflisch, A. | | Deposit date: | 2018-02-27 | | Release date: | 2018-10-10 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | In Silico Identification of JMJD3 Demethylase Inhibitors.
J Chem Inf Model, 58, 2018
|
|
5UTX
 
 | | Crystal structure of thioredoxin-disulfide reductase from Vibrio vulnificus CMCP6 - apo form | | Descriptor: | PHOSPHATE ION, Thioredoxin reductase | | Authors: | Chang, C, Grimshaw, S, Maltseva, N, Mulligan, R, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2017-02-15 | | Release date: | 2017-02-22 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.46 Å) | | Cite: | Crystal structure of thioredoxin-disulfide reductase from Vibrio vulnificus CMCP6 - apo form
To Be Published
|
|
8UTX
 
 | | Solution structure of a 12-mer peptide bearing a bicyclic Asx motif mimic (BAMM) as a synthetic N-cap | | Descriptor: | 1,3,5-tris(bromomethyl)benzene, TRP-CYS-ASP-ALA-ALA-CYS-CYS-ALA-ALA-ALA-LYS-ALA-NH2 peptide | | Authors: | Mi, T.X, Burgess, K. | | Deposit date: | 2023-10-31 | | Release date: | 2024-06-05 | | Last modified: | 2024-10-23 | | Method: | SOLUTION NMR | | Cite: | Bioinformatics leading to conveniently accessible, helix enforcing, bicyclic ASX motif mimics (BAMMs).
Nat Commun, 15, 2024
|
|
3UTX
 
 | |
6UTX
 
 | | E. coli sigma-S transcription initiation complex with an empty bubble ("Old" crystal) | | Descriptor: | DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, DNA-directed RNA polymerase subunit beta', ... | | Authors: | Zuo, Y, De, S, Steitz, T.A. | | Deposit date: | 2019-10-30 | | Release date: | 2020-08-26 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (4.05 Å) | | Cite: | Structural Insights into Transcription Initiation from De Novo RNA Synthesis to Transitioning into Elongation.
Iscience, 23, 2020
|
|
4UTX
 
 | | Crystal structure of zebrafish Sirtuin 5 in complex with 3-nitro- propionylated CPS1-peptide | | Descriptor: | 1,2-ETHANEDIOL, 3-NITROPROPANOIC ACID, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ... | | Authors: | Pannek, M, Gertz, M, Steegborn, C. | | Deposit date: | 2014-07-23 | | Release date: | 2014-08-20 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Chemical Probing of the Human Sirtuin 5 Active Site Reveals its Substrate Acyl Specificity and Peptide-Based Inhibitors.
Angew.Chem.Int.Ed.Engl., 53, 2014
|
|
1UTX
 
 | | Regulation of Cytolysin Expression by Enterococcus faecalis: Role of CylR2 | | Descriptor: | CYLR2, IODIDE ION, SODIUM ION | | Authors: | Razeto, A, Rumpel, S, Pillar, C.M, Gilmore, M.S, Becker, S, Zweckstetter, M. | | Deposit date: | 2003-12-12 | | Release date: | 2004-09-16 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure and DNA-Binding Properties of the Cytolysin Regulator CylR2 from Enterococcus Faecalis
Embo J., 23, 2004
|
|
2GZU
 
 | |
2XI8
 
 | | High resolution structure of native CylR2 | | Descriptor: | GLYCEROL, PUTATIVE TRANSCRIPTION REGULATOR | | Authors: | Gruene, T, Cho, M.-K, Karyagina, I, Kim, H.-Y, Grosse, C, Giller, K, Zweckstetter, M, Becker, S. | | Deposit date: | 2010-06-28 | | Release date: | 2011-02-09 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.21 Å) | | Cite: | Integrated Analysis of the Conformation of a Protein-Linked Spin Label by Crystallography, Epr and NMR Spectroscopy.
J.Biomol.NMR, 49, 2011
|
|
2XIU
 
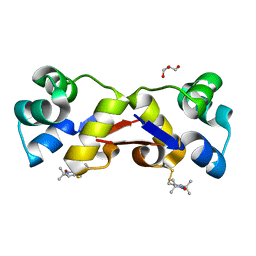 | | High resolution structure of MTSL-tagged CylR2. | | Descriptor: | CYLR2, GLYCEROL, S-[(1-oxyl-2,2,5,5-tetramethyl-2,5-dihydro-1H-pyrrol-3-yl)methyl] methanesulfonothioate | | Authors: | Gruene, T, Cho, M.-K, Karyagina, I, Kim, H.-Y, Grosse, C, Giller, K, Zweckstetter, M, Becker, S. | | Deposit date: | 2010-07-01 | | Release date: | 2011-02-09 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Integrated Analysis of the Conformation of a Protein-Linked Spin Label by Crystallography, Epr and NMR Spectroscopy.
J.Biomol.NMR, 49, 2011
|
|
2XJ3
 
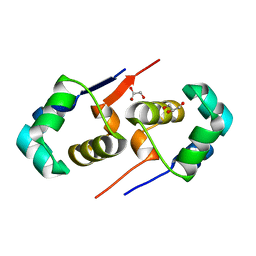 | | High resolution structure of the T55C mutant of CylR2. | | Descriptor: | CYLR2 SYNONYM CYTOLYSIN REPRESSOR 2, GLYCEROL | | Authors: | Gruene, T, Cho, M.K, Karyagina, I, Kim, H.Y, Grosse, C, Giller, K, Zweckstetter, M, Becker, S. | | Deposit date: | 2010-07-02 | | Release date: | 2011-02-09 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.23 Å) | | Cite: | Integrated Analysis of the Conformation of a Protein-Linked Spin Label by Crystallography, Epr and NMR Spectroscopy.
J.Biomol.NMR, 49, 2011
|
|
2LYR
 
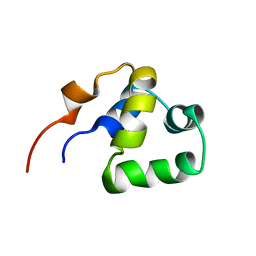 | | NOE-based 3D structure of the monomeric partially-folded intermediate of CylR2 at 259K (-14 Celsius degrees) | | Descriptor: | CylR2 | | Authors: | Jaremko, M, Jaremko, L, Kim, H, Cho, M, Schwieters, C.D, Giller, K, Becker, S, Zweckstetter, M. | | Deposit date: | 2012-09-19 | | Release date: | 2013-02-20 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Cold denaturation of a protein dimer monitored at atomic resolution.
Nat.Chem.Biol., 9, 2013
|
|
2LYP
 
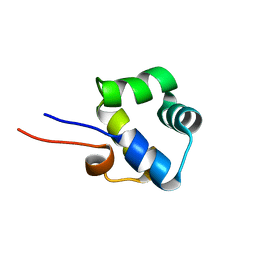 | | NOE-based 3D structure of the monomer of CylR2 in equilibrium with predissociated homodimer at 266K (-7 Celsius degrees) | | Descriptor: | CylR2 | | Authors: | Jaremko, M, Jaremko, L, Kim, H, Cho, M, Schwieters, C.D, Giller, K, Becker, S, Zweckstetter, M. | | Deposit date: | 2012-09-19 | | Release date: | 2013-02-20 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Cold denaturation of a protein dimer monitored at atomic resolution.
Nat.Chem.Biol., 9, 2013
|
|
2LYJ
 
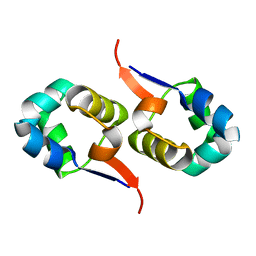 | | NOE-based 3D structure of the CylR2 homodimer at 298K | | Descriptor: | CylR2 | | Authors: | Jaremko, M, Jaremko, L, Kim, H, Cho, M, Giller, K, Becker, S, Zweckstetter, M, Schwieters, C.D. | | Deposit date: | 2012-09-19 | | Release date: | 2013-02-20 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Cold denaturation of a protein dimer monitored at atomic resolution.
Nat.Chem.Biol., 9, 2013
|
|
2LYK
 
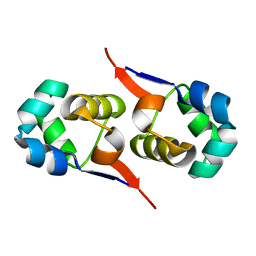 | | NOE-based 3D structure of the CylR2 homodimer at 270K (-3 Celsius degrees) | | Descriptor: | CylR2 | | Authors: | Jaremko, M, Jaremko, L, Kim, H, Cho, M, Schwieters, C.D, Giller, K, Becker, S, Zweckstetter, M. | | Deposit date: | 2012-09-19 | | Release date: | 2013-02-20 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Cold denaturation of a protein dimer monitored at atomic resolution.
Nat.Chem.Biol., 9, 2013
|
|
2LYQ
 
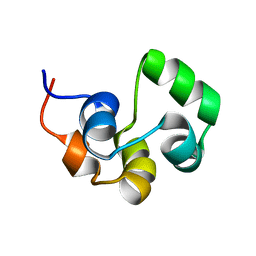 | | NOE-based 3D structure of the monomeric intermediate of CylR2 at 262K (-11 Celsius degrees) | | Descriptor: | CylR2 | | Authors: | Jaremko, M, Jaremko, L, Kim, H, Cho, M, Schwieters, C.D, Giller, K, Becker, S, Zweckstetter, M. | | Deposit date: | 2012-09-19 | | Release date: | 2013-02-20 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Cold denaturation of a protein dimer monitored at atomic resolution.
Nat.Chem.Biol., 9, 2013
|
|
2LYL
 
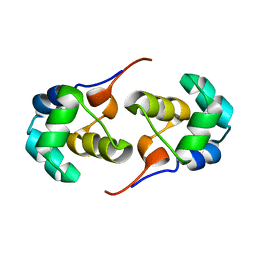 | | NOE-based 3D structure of the predissociated homodimer of CylR2 in equilibrium with monomer at 266K (-7 Celsius degrees) | | Descriptor: | CylR2 | | Authors: | Jaremko, M, Jaremko, L, Kim, H, Cho, M, Schwieters, C.D, Giller, K, Becker, S, Zweckstetter, M. | | Deposit date: | 2012-09-19 | | Release date: | 2013-02-20 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Cold denaturation of a protein dimer monitored at atomic resolution.
Nat.Chem.Biol., 9, 2013
|
|
