3BI9
 
 | | Tim-4 | | Descriptor: | ACETATE ION, T-cell immunoglobulin and mucin domain-containing protein 4 | | Authors: | Santiago, C, Ballesteros, A, Kaplan, G.G, Freeman, G.J, Casasnovas, J.M. | | Deposit date: | 2007-11-30 | | Release date: | 2008-01-01 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Structures of T Cell Immunoglobulin Mucin Protein 4 Show a Metal-Ion-Dependent Ligand Binding Site where Phosphatidylserine Binds.
Immunity, 27, 2007
|
|
3BIA
 
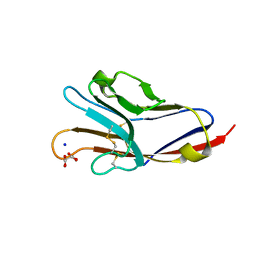 | | Tim-4 in complex with sodium potassium tartrate | | Descriptor: | L(+)-TARTARIC ACID, SODIUM ION, T-cell immunoglobulin and mucin domain-containing protein 4 | | Authors: | Santiago, C, Ballesteros, A, Kaplan, G.G, Freeman, G.J, Casasnovas, J.M. | | Deposit date: | 2007-11-30 | | Release date: | 2008-01-01 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structures of T Cell Immunoglobulin Mucin Protein 4 Show a Metal-Ion-Dependent Ligand Binding Site where Phosphatidylserine Binds.
Immunity, 27, 2007
|
|
3BIB
 
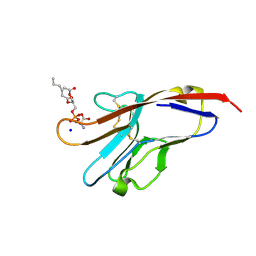 | | Tim-4 in complex with phosphatidylserine | | Descriptor: | 1,2-DICAPROYL-SN-PHOSPHATIDYL-L-SERINE, SODIUM ION, T-cell immunoglobulin and mucin domain-containing protein 4 | | Authors: | Santiago, C, Ballesteros, A, Kaplan, G.G, Freeman, G.J, Casasnovas, J.M. | | Deposit date: | 2007-11-30 | | Release date: | 2008-01-01 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structures of T Cell Immunoglobulin Mucin Protein 4 Show a Metal-Ion-Dependent Ligand Binding Site where Phosphatidylserine Binds.
Immunity, 27, 2007
|
|
3BIK
 
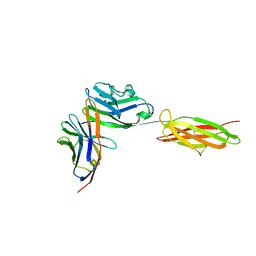 | | Crystal Structure of the PD-1/PD-L1 Complex | | Descriptor: | GLYCEROL, Programmed cell death 1 ligand 1, Programmed cell death protein 1 | | Authors: | Lin, D.Y, Tanaka, Y, Iwasaki, M, Gittis, A.G, Su, H.P, Mikami, B, Okazaki, T, Honjo, T, Minato, N, Garboczi, D.N. | | Deposit date: | 2007-11-30 | | Release date: | 2008-02-26 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | The PD-1/PD-L1 complex resembles the antigen-binding Fv domains of antibodies and T cell receptors.
Proc.Natl.Acad.Sci.Usa, 105, 2008
|
|
3BIS
 
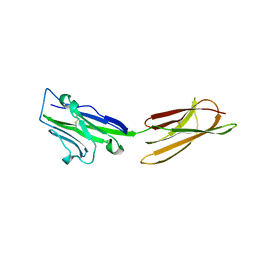 | | Crystal Structure of the PD-L1 | | Descriptor: | Programmed cell death 1 ligand 1 | | Authors: | Lin, D.Y, Tanaka, Y, Iwasaki, M, Gittis, A.G, Su, H.P, Mikami, B, Okazaki, T, Honjo, T, Minato, N, Garboczi, D.N. | | Deposit date: | 2007-11-30 | | Release date: | 2008-02-26 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.64 Å) | | Cite: | The PD-1/PD-L1 complex resembles the antigen-binding Fv domains of antibodies and T cell receptors.
Proc.Natl.Acad.Sci.Usa, 105, 2008
|
|
3BJ9
 
 | | Crystal structure of the Surrogate Light Chain Variable Domain VpreBJ | | Descriptor: | 1,2-ETHANEDIOL, Immunoglobulin iota chain, Immunoglobulin lambda-like polypeptide 1 | | Authors: | Morstadt, L.M, Bohm, A.A, Stollar, B.D, Baleja, J.D. | | Deposit date: | 2007-12-03 | | Release date: | 2008-03-04 | | Last modified: | 2019-09-04 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Engineering and characterization of a single chain surrogate light chain variable domain.
Protein Sci., 17, 2008
|
|
3BKC
 
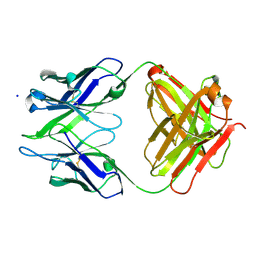 | | Crystal structure of anti-amyloid beta FAB WO2 (P21, FormB) | | Descriptor: | SODIUM ION, WO2 IgG2a Fab fragment Heavy Chain, WO2 IgG2a Fab fragment Light Chain Kappa | | Authors: | Miles, L.A, Wun, K.S, Crespi, G.A, Fodero-Tavoletti, M, Galatis, D, Bageley, C.J, Beyreuther, K, Masters, C.L, Cappai, R, McKinstry, W.J, Barnham, K.J, Parker, M.W. | | Deposit date: | 2007-12-06 | | Release date: | 2008-04-15 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Amyloid-beta-anti-amyloid-beta complex structure reveals an extended conformation in the immunodominant B-cell epitope.
J.Mol.Biol., 377, 2008
|
|
3BKJ
 
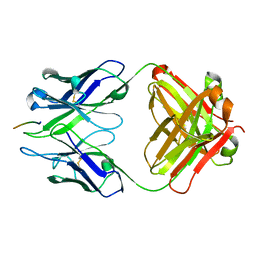 | | Crystal structure of Fab wo2 bound to the n terminal domain of amyloid beta peptide (1-16) | | Descriptor: | Amyloid Beta Peptide, WO2 IgG2a Fab fragment Heavy Chain, WO2 IgG2a Fab fragment Light Chain Kappa | | Authors: | Miles, L.A, Wun, K.S, Crespi, G.A, Fodero-Tavoletti, M, Galatis, D, Bageley, C.J, Beyreuther, K, Masters, C.L, Cappai, R, McKinstry, W.J, Barnham, K.J, Parker, M.W. | | Deposit date: | 2007-12-06 | | Release date: | 2008-04-15 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Amyloid-beta-anti-amyloid-beta complex structure reveals an extended conformation in the immunodominant B-cell epitope.
J.Mol.Biol., 377, 2008
|
|
3BKM
 
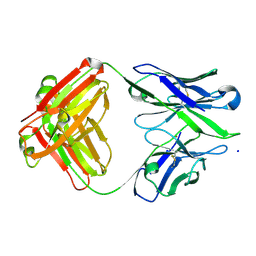 | | Structure of anti-amyloid-beta Fab WO2 (Form A, P212121) | | Descriptor: | SODIUM ION, WO2 IgG2a Fab fragment Heavy Chain, WO2 IgG2a Fab fragment Light Chain Kappa, ... | | Authors: | Miles, L.A, Wun, K.S, Crespi, G.A, Fodero-Tavoletti, M, Galatis, D, Bageley, C.J, Beyreuther, K, Masters, C.L, Cappai, R, McKinstry, W.J, Barnham, K.J, Parker, M.W. | | Deposit date: | 2007-12-07 | | Release date: | 2008-04-15 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Amyloid-beta-anti-amyloid-beta complex structure reveals an extended conformation in the immunodominant B-cell epitope.
J.Mol.Biol., 377, 2008
|
|
3BN9
 
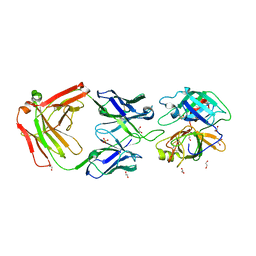 | | Crystal Structure of MT-SP1 in complex with Fab Inhibitor E2 | | Descriptor: | 1,2-ETHANEDIOL, E2 Fab Heavy Chain, E2 Fab Light Chain, ... | | Authors: | Farady, C.J, Schneider, E.L, Egea, P.F, Goetz, D.H, Craik, C.S. | | Deposit date: | 2007-12-13 | | Release date: | 2008-09-09 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.173 Å) | | Cite: | Structure of an Fab-protease complex reveals a highly specific non-canonical mechanism of inhibition
J.Mol.Biol., 380, 2008
|
|
3BP5
 
 | | Crystal structure of the mouse PD-1 and PD-L2 complex | | Descriptor: | GLYCEROL, Programmed cell death 1 ligand 2, Programmed cell death protein 1 | | Authors: | Yan, Q, Lazar-Molnar, E, Cao, E, Ramagopal, U.A, Toro, R, Nathenson, S.G, Almo, S.C. | | Deposit date: | 2007-12-18 | | Release date: | 2008-07-15 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of the complex between programmed death-1 (PD-1) and its ligand PD-L2.
Proc.Natl.Acad.Sci.Usa, 105, 2008
|
|
3BP6
 
 | | Crystal structure of the mouse PD-1 Mutant and PD-L2 complex | | Descriptor: | GLYCEROL, Programmed cell death 1 ligand 2, Programmed cell death protein 1 | | Authors: | Yan, Q, Lazar-Molnar, E, Cao, E, Ramagopal, U.A, Toro, R, Nathenson, S.G, Almo, S.C. | | Deposit date: | 2007-12-18 | | Release date: | 2009-02-24 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structure of the mouse PD-1 A99L and PD-L2 complex
To be published
|
|
3BQU
 
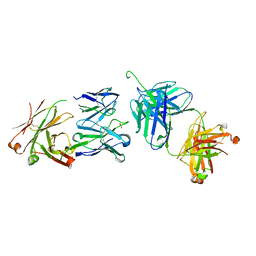 | | Crystal Structure of the 2F5 Fab'-3H6 Fab Complex | | Descriptor: | 2F5 Fab' heavy chain, 2F5 Fab' light chain, 3H6 Fab heavy chain, ... | | Authors: | Bryson, S, Julien, J.-P, Isenman, D.E, Kunert, R, Katinger, H, Pai, E.F. | | Deposit date: | 2007-12-20 | | Release date: | 2008-10-21 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal structure of the complex between the F(ab)' fragment of the cross-neutralizing anti-HIV-1 antibody 2F5 and the F(ab) fragment of its anti-idiotypic antibody 3H6.
J.Mol.Biol., 382, 2008
|
|
3BX7
 
 | |
3CDC
 
 | |
3CDF
 
 | |
3CDY
 
 | | AL-09 H87Y, immunoglobulin light chain variable domain | | Descriptor: | IMMUNOGLOBULIN LIGHT CHAIN | | Authors: | Baden, E.M, Randles, E.G, Aboagye, A.K, Thompson, J.R, Ramirez-Alvarado, M. | | Deposit date: | 2008-02-27 | | Release date: | 2008-09-02 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.43 Å) | | Cite: | Structural insights into the role of mutations in amyloidogenesis.
J.Biol.Chem., 283, 2008
|
|
3CFJ
 
 | |
3CFK
 
 | | Crystal structure of catalytic elimination antibody 34E4, triclinic crystal form | | Descriptor: | 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CADMIUM ION, CATALYTIC ANTIBODY FAB 34E4 HEAVY CHAIN,Uncharacterized protein, ... | | Authors: | Debler, E.W, Wilson, I.A. | | Deposit date: | 2008-03-04 | | Release date: | 2008-04-15 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Conformational isomerism can limit antibody catalysis.
J.Biol.Chem., 283, 2008
|
|
3CHN
 
 | | Solution structure of human secretory IgA1 | | Descriptor: | Ig alpha-1 chain C region, Immunoglobulin kappa light chain, Secretory component | | Authors: | Bonner, A, Almogren, A, Furtado, P.B, Kerr, M.A, Perkins, S.J. | | Deposit date: | 2008-03-10 | | Release date: | 2008-12-30 | | Last modified: | 2024-02-21 | | Method: | SOLUTION SCATTERING | | Cite: | Location of secretory component on the Fc edge of dimeric IgA1 reveals insight into the role of secretory IgA1 in mucosal immunity.
Mucosal Immunol, 2, 2009
|
|
3CM9
 
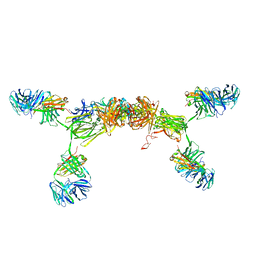 | | Solution Structure of Human SIgA2 | | Descriptor: | Immunoglobulin heavy chain, Immunoglobulin light chain, Secretory component | | Authors: | Bonner, A, Almogren, A, Furtado, P.B, Kerr, M.A, Perkins, S.J. | | Deposit date: | 2008-03-21 | | Release date: | 2008-12-30 | | Last modified: | 2024-02-21 | | Method: | SOLUTION SCATTERING | | Cite: | The nonplanar secretory IgA2 and near planar secretory IgA1 solution structures rationalize their different mucosal immune responses.
J.Biol.Chem., 284, 2009
|
|
3CMO
 
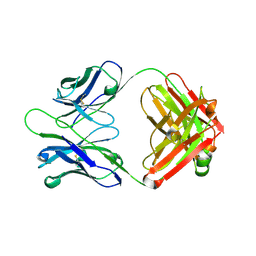 | |
3CSP
 
 | | Crystal structure of the DM2 mutant of myelin oligodendrocyte glycoprotein | | Descriptor: | Myelin-oligodendrocyte glycoprotein | | Authors: | Breithaupt, C, Schafer, B, Pellkofer, H, Huber, R, Linington, C, Jacob, U. | | Deposit date: | 2008-04-10 | | Release date: | 2008-10-21 | | Last modified: | 2021-10-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Demyelinating myelin oligodendrocyte glycoprotein-specific autoantibody response is focused on one dominant conformational epitope region in rodents
J.Immunol., 181, 2008
|
|
3D0L
 
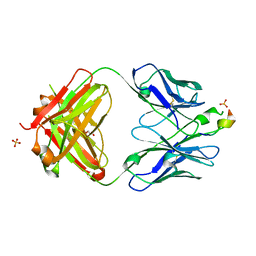 | | Crystal structure of the HIV-1 broadly neutralizing antibody 2F5 in complex with the gp41 FP-MPER Hyb3K construct 514GIGALFLGFLGAAGS528KK-Ahx-655KNEQELLELDKWASLWN671 | | Descriptor: | 2F5 heavy chain, 2F5 light chain, GLYCEROL, ... | | Authors: | Bryson, S, Julien, J.P, Pai, E.F. | | Deposit date: | 2008-05-01 | | Release date: | 2008-10-07 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structural details of HIV-1 recognition by the broadly neutralizing monoclonal antibody 2F5: epitope conformation, antigen-recognition loop mobility, and anion-binding site.
J.Mol.Biol., 384, 2008
|
|
3DMM
 
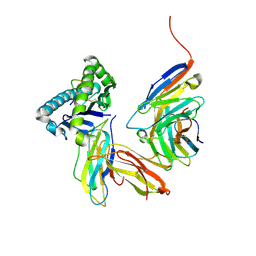 | | Crystal structure of the CD8 alpha beta/H-2Dd complex | | Descriptor: | Beta-2 microglobulin, H-2 class I histocompatibility antigen, D-D alpha chain, ... | | Authors: | Wang, R, Natarajan, K, Margulies, D.H. | | Deposit date: | 2008-07-01 | | Release date: | 2009-07-14 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural basis of the CD8alphabeta/MHC class i interaction: focused recognition orients CD8beta to a T cell proximal position.
J.Immunol., 183, 2009
|
|
