6RKS
 
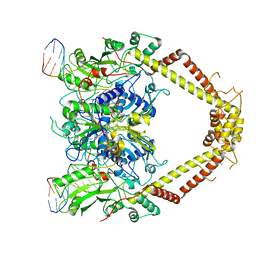 | | E. coli DNA Gyrase - DNA binding and cleavage domain in State 1 without TOPRIM insertion | | Descriptor: | (3~{R})-3-[[4-(3,4-dihydro-2~{H}-pyrano[2,3-c]pyridin-6-ylmethylamino)piperidin-1-yl]methyl]-1,4,7-triazatricyclo[6.3.1.0^{4,12}]dodeca-6,8(12),9-triene-5,11-dione, DNA Strand 1, DNA Strand 2, ... | | Authors: | Vanden Broeck, A, Lamour, V. | | Deposit date: | 2019-04-30 | | Release date: | 2019-11-06 | | Last modified: | 2020-11-18 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Cryo-EM structure of the complete E. coli DNA gyrase nucleoprotein complex.
Nat Commun, 10, 2019
|
|
6RKV
 
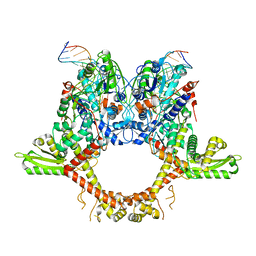 | |
6RKU
 
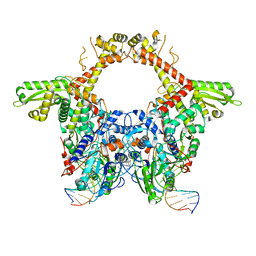 | | E. coli DNA Gyrase - DNA binding and cleavage domain in State 1 | | Descriptor: | (3~{R})-3-[[4-(3,4-dihydro-2~{H}-pyrano[2,3-c]pyridin-6-ylmethylamino)piperidin-1-yl]methyl]-1,4,7-triazatricyclo[6.3.1.0^{4,12}]dodeca-6,8(12),9-triene-5,11-dione, DNA Strand 1, DNA Strand 2, ... | | Authors: | Vanden Broeck, A, Lamour, V. | | Deposit date: | 2019-04-30 | | Release date: | 2019-11-06 | | Last modified: | 2020-11-18 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Cryo-EM structure of the complete E. coli DNA gyrase nucleoprotein complex.
Nat Commun, 10, 2019
|
|
6QRJ
 
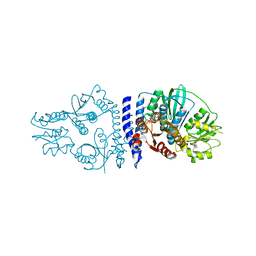 | |
6S1K
 
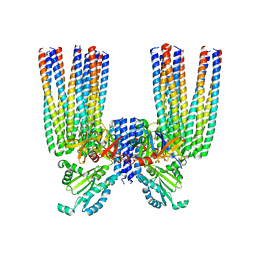 | | E. coli Core Signaling Unit, carrying QQQQ receptor mutation | | Descriptor: | CheW, Chemotaxis protein CheA, Methyl-accepting chemotaxis protein I | | Authors: | Cassidy, C.K. | | Deposit date: | 2019-06-18 | | Release date: | 2020-01-22 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (8.38 Å) | | Cite: | Structure and dynamics of the E. coli chemotaxis core signaling complex by cryo-electron tomography and molecular simulations.
Commun Biol, 3, 2020
|
|
6J90
 
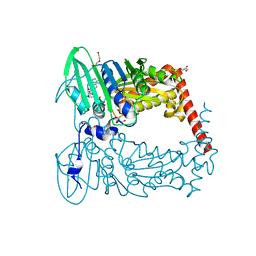 | | Crystal Structure of GyraseB N-Terminal Domain complex with ATP from Salmonella Typhi at 2.2A Resolution | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Kaur, G, Sachdeva, E, Tiwari, P, Gupta, D, Ethayathulla, A.S, Kaur, P. | | Deposit date: | 2019-01-21 | | Release date: | 2020-01-22 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal Structure of GyraseB N-Terminal Domain complex with ATP from Salmonella Typhi at 2.2A Resolution
To Be Published
|
|
6RGZ
 
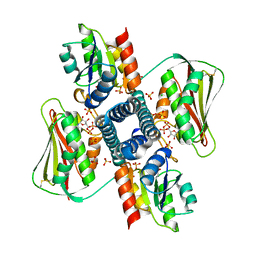 | | Revisiting pH-gated conformational switch. Complex HK853-RR468 pH 6.5 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, CHLORIDE ION, MAGNESIUM ION, ... | | Authors: | Mideros-Mora, C, Casino, P, Marina, A. | | Deposit date: | 2019-04-18 | | Release date: | 2020-02-19 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Revisiting the pH-gated conformational switch on the activities of HisKA-family histidine kinases.
Nat Commun, 11, 2020
|
|
6RH8
 
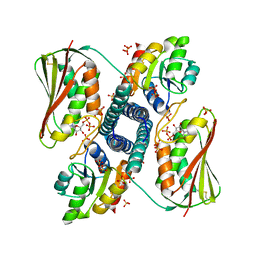 | | Revisiting pH-gated conformational switch. Complex HK853 mutant H260A -RR468 mutant D53A pH 5.3 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Response regulator, SULFATE ION, ... | | Authors: | Mideros-Mora, C, Casino, P, Marina, A. | | Deposit date: | 2019-04-19 | | Release date: | 2020-02-19 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Revisiting the pH-gated conformational switch on the activities of HisKA-family histidine kinases.
Nat Commun, 11, 2020
|
|
6RGY
 
 | | Revisiting pH-gated conformational switch. Complex HK853-RR468 pH 7.5 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, CITRIC ACID, MAGNESIUM ION, ... | | Authors: | Mideros-Mora, C, Casino, P, Marina, A. | | Deposit date: | 2019-04-18 | | Release date: | 2020-02-19 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Revisiting the pH-gated conformational switch on the activities of HisKA-family histidine kinases.
Nat Commun, 11, 2020
|
|
6RFV
 
 | | Revisiting pH-gated conformational switch. Complex HK853-RR468 pH 7 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, Response regulator, ... | | Authors: | Mideros-Mora, C, Casino, P, Marina, A. | | Deposit date: | 2019-04-16 | | Release date: | 2020-02-19 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.83 Å) | | Cite: | Revisiting the pH-gated conformational switch on the activities of HisKA-family histidine kinases.
Nat Commun, 11, 2020
|
|
6RH2
 
 | | Revisiting pH-gated conformational switch. Complex HK853-RR468 D53A pH 5.3 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Response regulator, SULFATE ION, ... | | Authors: | Mideros-Mora, C, Casino, P, Marina, A. | | Deposit date: | 2019-04-18 | | Release date: | 2020-02-19 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Revisiting the pH-gated conformational switch on the activities of HisKA-family histidine kinases.
Nat Commun, 11, 2020
|
|
6RH7
 
 | | Revisiting pH-gated conformational switch. Complex HK853 mutant H260A -RR468 mutant D53A pH 7.5 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Response regulator, SULFATE ION, ... | | Authors: | Mideros-Mora, C, Casino, P, Marina, A. | | Deposit date: | 2019-04-18 | | Release date: | 2020-02-19 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Revisiting the pH-gated conformational switch on the activities of HisKA-family histidine kinases.
Nat Commun, 11, 2020
|
|
6RH1
 
 | | Revisiting pH-gated conformational switch. Complex HK853-RR468 D53A pH 7 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Response regulator, SULFATE ION, ... | | Authors: | Mideros-Mora, C, Casino, P, Marina, A. | | Deposit date: | 2019-04-18 | | Release date: | 2020-02-19 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Revisiting the pH-gated conformational switch on the activities of HisKA-family histidine kinases.
Nat Commun, 11, 2020
|
|
6RH0
 
 | | Revisiting pH-gated conformational switch. Complex HK853-RR468 pH 5.5 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, Response regulator, ... | | Authors: | Mideros-Mora, C, Casino, P, Marina, A. | | Deposit date: | 2019-04-18 | | Release date: | 2020-02-19 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.87 Å) | | Cite: | Revisiting the pH-gated conformational switch on the activities of HisKA-family histidine kinases.
Nat Commun, 11, 2020
|
|
6PAJ
 
 | |
6KZX
 
 | | Crystal structure of E.coli DNA gyrase B in complex with 2-oxo-1,2-dihydroquinoline derivative | | Descriptor: | 3-[[8-(methylamino)-2-oxidanylidene-1~{H}-quinolin-3-yl]carbonylamino]benzoic acid, DNA gyrase subunit B | | Authors: | Mima, M, Takeuchi, T, Ushiyama, F. | | Deposit date: | 2019-09-25 | | Release date: | 2020-05-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Lead Identification of 8-(Methylamino)-2-oxo-1,2-dihydroquinoline Derivatives as DNA Gyrase Inhibitors: Hit-to-Lead Generation Involving Thermodynamic Evaluation.
Acs Omega, 5, 2020
|
|
6L01
 
 | | Crystal structure of E.coli DNA gyrase B in complex with 2-oxo-1,2-dihydroquinoline derivative | | Descriptor: | 2-[3-[[8-(methylamino)-2-oxidanylidene-1~{H}-quinolin-3-yl]carbonylamino]phenyl]ethanoic acid, DNA gyrase subunit B | | Authors: | Mima, M, Takeuchi, T, Ushiyama, F. | | Deposit date: | 2019-09-25 | | Release date: | 2020-05-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Lead Identification of 8-(Methylamino)-2-oxo-1,2-dihydroquinoline Derivatives as DNA Gyrase Inhibitors: Hit-to-Lead Generation Involving Thermodynamic Evaluation.
Acs Omega, 5, 2020
|
|
6KZV
 
 | |
6KZZ
 
 | | Crystal structure of E.coli DNA gyrase B in complex with 2-oxo-1,2-dihydroquinoline derivative | | Descriptor: | 4-[[8-(methylamino)-2-oxidanylidene-1~{H}-quinolin-3-yl]carbonylamino]benzoic acid, DNA gyrase subunit B | | Authors: | Mima, M, Takeuchi, T, Ushiyama, F. | | Deposit date: | 2019-09-25 | | Release date: | 2020-05-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Lead Identification of 8-(Methylamino)-2-oxo-1,2-dihydroquinoline Derivatives as DNA Gyrase Inhibitors: Hit-to-Lead Generation Involving Thermodynamic Evaluation.
Acs Omega, 5, 2020
|
|
6Y8O
 
 | | Mycobacterium smegmatis GyrB 22kDa ATPase sub-domain in complex with novobiocin | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, DNA gyrase subunit B, ... | | Authors: | Henderson, S.R, Stevenson, C.E.M, Malone, B, Zholnerovych, Y, Mitchenall, L.A, Pichowicz, M, McGarry, D.H, Cooper, I.R, Charrier, C, Salisbury, A, Lawson, D.M, Maxwell, A. | | Deposit date: | 2020-03-05 | | Release date: | 2020-08-12 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural and mechanistic analysis of ATPase inhibitors targeting mycobacterial DNA gyrase.
J.Antimicrob.Chemother., 75, 2020
|
|
6Y8L
 
 | | Mycobacterium thermoresistibile GyrB21 in complex with novobiocin | | Descriptor: | 1,2-ETHANEDIOL, DNA gyrase subunit B, NOVOBIOCIN, ... | | Authors: | Henderson, S.R, Stevenson, C.E.M, Malone, B, Zholnerovych, Y, Mitchenall, L.A, Pichowicz, M, McGarry, D.H, Cooper, I.R, Charrier, C, Salisbury, A, Lawson, D.M, Maxwell, A. | | Deposit date: | 2020-03-05 | | Release date: | 2020-08-12 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structural and mechanistic analysis of ATPase inhibitors targeting mycobacterial DNA gyrase.
J.Antimicrob.Chemother., 75, 2020
|
|
6Y8N
 
 | | Mycobacterium thermoresistibile GyrB21 in complex with Redx03863 | | Descriptor: | 1,2-ETHANEDIOL, 4-[(1~{S},5~{R})-6-azanyl-3-azabicyclo[3.1.0]hexan-3-yl]-6-fluoranyl-~{N}-methyl-2-(2-methylpyrimidin-5-yl)oxy-9~{H}-pyrimido[4,5-b]indol-8-amine, DNA gyrase subunit B, ... | | Authors: | Henderson, S.R, Stevenson, C.E.M, Malone, B, Zholnerovych, Y, Mitchenall, L.A, Pichowicz, M, McGarry, D.H, Cooper, I.R, Charrier, C, Salisbury, A, Lawson, D.M, Maxwell, A. | | Deposit date: | 2020-03-05 | | Release date: | 2020-08-12 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural and mechanistic analysis of ATPase inhibitors targeting mycobacterial DNA gyrase.
J.Antimicrob.Chemother., 75, 2020
|
|
6M1J
 
 | | The DNA Gyrase B ATP binding domain of PSEUDOMONAS AERUGINOSA in complex with compound 12x | | Descriptor: | 1-[5-[6-fluoranyl-8-(methylamino)-4-[3-(trifluoromethyl)pyrazol-1-yl]-9H-pyrido[2,3-b]indol-3-yl]pyrimidin-2-yl]cyclopropane-1-carboxylic acid, DIMETHYL SULFOXIDE, DNA gyrase subunit B, ... | | Authors: | Xu, Z.H, Zhou, Z. | | Deposit date: | 2020-02-26 | | Release date: | 2020-09-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.701 Å) | | Cite: | Discovery of Pyrido[2,3-b]indole Derivatives with Gram-Negative Activity Targeting Both DNA Gyrase and Topoisomerase IV.
J.Med.Chem., 63, 2020
|
|
6M1S
 
 | | The DNA Gyrase B ATP binding domain of PSEUDOMONAS AERUGINOSA in complex with compound 12o | | Descriptor: | 3-[5-[8-(ethylamino)-6-fluoranyl-4-[3-(trifluoromethyl)pyrazol-1-yl]-9H-pyrido[2,3-b]indol-3-yl]pyrimidin-2-yl]oxy-2,2-dimethyl-propanoic acid, CHLORIDE ION, DNA gyrase subunit B, ... | | Authors: | Xu, Z.H, Zhou, Z. | | Deposit date: | 2020-02-26 | | Release date: | 2020-09-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.254 Å) | | Cite: | Discovery of Pyrido[2,3-b]indole Derivatives with Gram-Negative Activity Targeting Both DNA Gyrase and Topoisomerase IV.
J.Med.Chem., 63, 2020
|
|
6TCK
 
 | | Crystal structure of the ATP binding domain of S. aureus GyrB complexed with ULD-2 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 2-[[3,4-bis(chloranyl)-5-methyl-1~{H}-pyrrol-2-yl]carbonylamino]-4-phenylmethoxy-1,3-benzothiazole-6-carboxylic acid, CALCIUM ION, ... | | Authors: | Welin, M, Kimbung, R, Focht, D. | | Deposit date: | 2019-11-06 | | Release date: | 2020-09-23 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Rational design of balanced dual-targeting antibiotics with limited resistance.
Plos Biol., 18, 2020
|
|
