7TE2
 
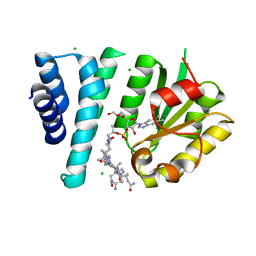 | | Crystal Structure of AerR from Rhodobacter capsulatus at 2.25 A. | | Descriptor: | AerR, CHLORIDE ION, COBALAMIN, ... | | Authors: | Dragnea, V, Gonzalez-Gutierrez, G, Bauer, C.E. | | Deposit date: | 2022-01-04 | | Release date: | 2022-08-31 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structural Analyses of CrtJ and Its B 12 -Binding Co-Regulators SAerR and LAerR from the Purple Photosynthetic Bacterium Rhodobacter capsulatus.
Microorganisms, 10, 2022
|
|
7KDY
 
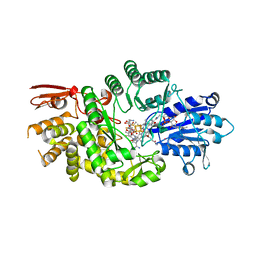 | | Crystal structure of Streptomyces tokunonesis TokK with hydroxycobalamin, 5'-deoxyadenosine, methionine, and (2R)-pantetheinylated carbapenam | | Descriptor: | (2R,3R,5R)-3-{[2-({N-[(2R)-2,4-dihydroxy-3,3-dimethylbutanoyl]-beta-alanyl}amino)ethyl]sulfanyl}-7-oxo-1-azabicyclo[3.2.0]heptane-2-carboxylic acid, 5'-DEOXYADENOSINE, COBALAMIN, ... | | Authors: | Knox, H.L, Booker, S.J, Boal, A.K. | | Deposit date: | 2020-10-09 | | Release date: | 2022-01-12 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.939 Å) | | Cite: | Structure of a B 12 -dependent radical SAM enzyme in carbapenem biosynthesis.
Nature, 602, 2022
|
|
7KDX
 
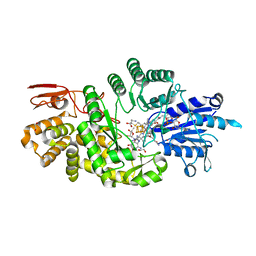 | | Crystal structure of Streptomyces tokunonesis TokK with hydroxycobalamin, 5'-deoxyadenosine, and methionine | | Descriptor: | 1,2-ETHANEDIOL, 5'-DEOXYADENOSINE, CHLORIDE ION, ... | | Authors: | Knox, H.L, Booker, S.J, Boal, A.K. | | Deposit date: | 2020-10-09 | | Release date: | 2022-01-12 | | Last modified: | 2022-02-23 | | Method: | X-RAY DIFFRACTION (1.791 Å) | | Cite: | Structure of a B 12 -dependent radical SAM enzyme in carbapenem biosynthesis.
Nature, 602, 2022
|
|
4Q37
 
 | |
1FMF
 
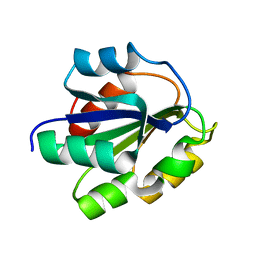 | | REFINED SOLUTION STRUCTURE OF THE (13C,15N-LABELED) B12-BINDING SUBUNIT OF GLUTAMATE MUTASE FROM CLOSTRIDIUM TETANOMORPHUM | | Descriptor: | METHYLASPARTATE MUTASE S CHAIN | | Authors: | Hoffmann, B, Konrat, R, Tollinger, M, Huhta, M, Marsh, E.N.G, Kraeutler, B. | | Deposit date: | 2000-08-17 | | Release date: | 2002-02-15 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | A protein pre-organized to trap the nucleotide moiety of coenzyme B(12): refined solution structure of the B(12)-binding subunit of glutamate mutase from Clostridium tetanomorphum.
Chembiochem, 2, 2001
|
|
1ID8
 
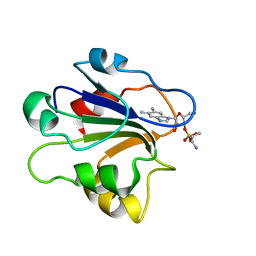 | | NMR STRUCTURE OF GLUTAMATE MUTASE (B12-BINDING SUBUNIT) COMPLEXED WITH THE VITAMIN B12 NUCLEOTIDE | | Descriptor: | 2-HYDROXY-PROPYL-AMMONIUM, METHYLASPARTATE MUTASE S CHAIN, PHOSPHORIC ACID MONO-[5-(5,6-DIMETHYL-BENZOIMIDAZOL-1-YL)-4-HYDROXY-2-HYDROXYMETHYL-TETRAHYDRO-FURAN-3-YL] ESTER | | Authors: | Tollinger, M, Eichmuller, C, Konrat, R, Huhta, M.S, Marsh, E.N.G, Krautler, B. | | Deposit date: | 2001-04-04 | | Release date: | 2001-06-27 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | The B(12)-binding subunit of glutamate mutase from Clostridium tetanomorphum traps the nucleotide moiety of coenzyme B(12).
J.Mol.Biol., 309, 2001
|
|
1B1A
 
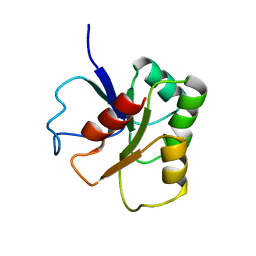 | | GLUTAMATE MUTASE (B12-BINDING SUBUNIT), NMR, MINIMIZED AVERAGE STRUCTURE | | Descriptor: | GLUTAMATE MUTASE | | Authors: | Hoffmann, B, Konrat, R, Bothe, H, Buckel, W, Kraeutler, B. | | Deposit date: | 1998-11-19 | | Release date: | 1999-07-13 | | Last modified: | 2024-04-10 | | Method: | SOLUTION NMR | | Cite: | Structure and dynamics of the B12-binding subunit of glutamate mutase from Clostridium cochlearium.
Eur.J.Biochem., 263, 1999
|
|
1BE1
 
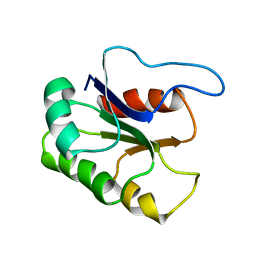 | | GLUTAMATE MUTASE (B12-BINDING SUBUNIT), NMR, MINIMIZED AVERAGE STRUCTURE | | Descriptor: | GLUTAMATE MUTASE | | Authors: | Tollinger, M, Konrat, R, Hilbert, B.H, Marsh, E.N.G, Kraeutler, B. | | Deposit date: | 1998-05-19 | | Release date: | 1998-08-26 | | Last modified: | 2022-02-16 | | Method: | SOLUTION NMR | | Cite: | How a protein prepares for B12 binding: structure and dynamics of the B12-binding subunit of glutamate mutase from Clostridium tetanomorphum
Structure, 6, 1998
|
|
2YXB
 
 | |
7XCN
 
 | | Crystal structure of the MttB-MttC complex at 2.7 A resolution | | Descriptor: | 5-HYDROXYBENZIMIDAZOLYLCOBAMIDE, GLYCEROL, Trimethylamine methyltransferase, ... | | Authors: | Li, J, Chan, M.K. | | Deposit date: | 2022-03-24 | | Release date: | 2023-01-18 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Insights into pyrrolysine function from structures of a trimethylamine methyltransferase and its corrinoid protein complex.
Commun Biol, 6, 2023
|
|
4JGI
 
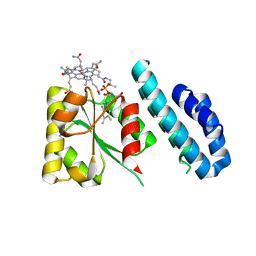 | | 1.5 Angstrom crystal structure of a novel cobalamin-binding protein from Desulfitobacterium hafniense DCB-2 | | Descriptor: | CO-METHYLCOBALAMIN, Putative uncharacterized protein | | Authors: | Sjuts, H, Dunstan, M.S, Fisher, K, Leys, D. | | Deposit date: | 2013-03-01 | | Release date: | 2013-08-07 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure of the cobalamin-binding protein of a putative O-demethylase from Desulfitobacterium hafniense DCB-2.
Acta Crystallogr.,Sect.D, 69, 2013
|
|
2I2X
 
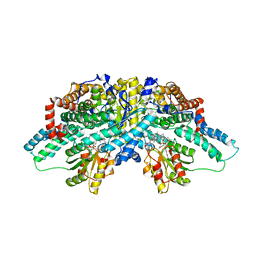 | | Crystal structure of methanol:cobalamin methyltransferase complex MtaBC from Methanosarcina barkeri | | Descriptor: | 5-HYDROXYBENZIMIDAZOLYLCOB(III)AMIDE, Methyltransferase 1, POTASSIUM ION, ... | | Authors: | Hagemeier, C.H, Kruer, M, Thauer, R.K, Warkentin, E, Ermler, U. | | Deposit date: | 2006-08-17 | | Release date: | 2006-11-21 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Insight into the mechanism of biological methanol activation based on the crystal structure of the methanol-cobalamin methyltransferase complex
Proc.Natl.Acad.Sci.Usa, 103, 2006
|
|
3EZX
 
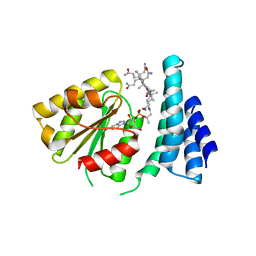 | |
8DPB
 
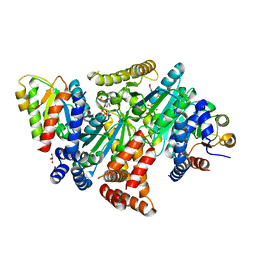 | | MeaB in complex with the cobalamin-binding domain of its target mutase with GMPPCP bound | | Descriptor: | GLYCEROL, MAGNESIUM ION, Methylmalonyl-CoA mutase accessory protein, ... | | Authors: | Vaccaro, F.A, Born, D.A, Drennan, C.L. | | Deposit date: | 2022-07-15 | | Release date: | 2023-03-01 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.72 Å) | | Cite: | Structure of metallochaperone in complex with the cobalamin-binding domain of its target mutase provides insight into cofactor delivery.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
6WTE
 
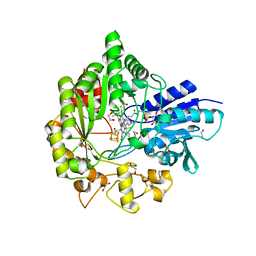 | | Structure of radical S-adenosylmethionine methyltransferase, TsrM, from Kitasatospora setae with cobalamin and [4Fe-4S] cluster bound | | Descriptor: | 1,2-ETHANEDIOL, B12-binding domain-containing protein, COBALAMIN, ... | | Authors: | Knox, H.L, Chen, P.Y.-T, Drennan, C.L, Booker, S.J. | | Deposit date: | 2020-05-02 | | Release date: | 2020-12-23 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Structural basis for non-radical catalysis by TsrM, a radical SAM methylase.
Nat.Chem.Biol., 17, 2021
|
|
6WTF
 
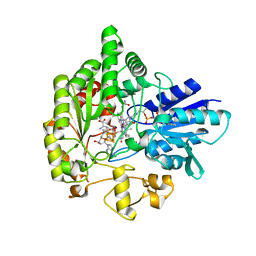 | | Structure of radical S-adenosylmethionine methyltransferase, TsrM, from Kitasatospora setae with tryptophan substrate and SAM analog (aza-SAM) bound | | Descriptor: | COBALAMIN, IRON/SULFUR CLUSTER, S-5'-AZAMETHIONINE-5'-DEOXYADENOSINE, ... | | Authors: | Knox, H.L, Chen, P.Y.-T, Drennan, C.L, Booker, S.J. | | Deposit date: | 2020-05-02 | | Release date: | 2020-12-23 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Structural basis for non-radical catalysis by TsrM, a radical SAM methylase.
Nat.Chem.Biol., 17, 2021
|
|
1Y80
 
 | | Structure of a corrinoid (factor IIIm)-binding protein from Moorella thermoacetica | | Descriptor: | CO-5-METHOXYBENZIMIDAZOLYLCOBAMIDE, Predicted cobalamin binding protein, UNKNOWN ATOM OR ION | | Authors: | Liu, Z.-J, Fu, Z.-Q, Tempel, W, Das, A, Habel, J, Zhou, W, Chang, J, Chen, L, Lee, D, Nguyen, D, Chang, S.-H, Ljungdahl, L, Rose, J.P, Wang, B.-C, Southeast Collaboratory for Structural Genomics (SECSG) | | Deposit date: | 2004-12-10 | | Release date: | 2005-01-25 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure of a corrinoid (factor IIIm)-binding protein from Moorella thermoacetica
To be published
|
|
6H9F
 
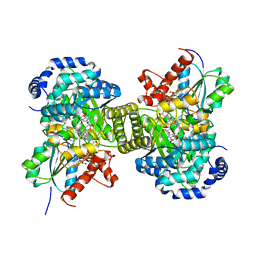 | | Structure of glutamate mutase reconstituted with bishomo-coenzyme B12 | | Descriptor: | (2~{R},3~{R},4~{S},5~{R})-2-(6-aminopurin-9-yl)-5-propyl-oxolane-3,4-diol, COBALAMIN, D(-)-TARTARIC ACID, ... | | Authors: | Gruber, K, Csitkovits, V, Kratky, C. | | Deposit date: | 2018-08-03 | | Release date: | 2019-08-14 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure-Based Demystification of Radical Catalysis by a Coenzyme B 12 Dependent Enzyme-Crystallographic Study of Glutamate Mutase with Cofactor Homologues.
Angew.Chem.Int.Ed.Engl., 61, 2022
|
|
6H9E
 
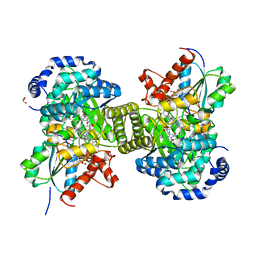 | | Structure of glutamate mutase reconstituted with homo-coenzyme B12 | | Descriptor: | (2~{R},3~{R},4~{S},5~{R})-2-(6-aminopurin-9-yl)-5-ethyl-oxolane-3,4-diol, COBALAMIN, D(-)-TARTARIC ACID, ... | | Authors: | Gruber, K, Csitkovits, V, Kratky, C. | | Deposit date: | 2018-08-03 | | Release date: | 2019-08-14 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Structure-Based Demystification of Radical Catalysis by a Coenzyme B 12 Dependent Enzyme-Crystallographic Study of Glutamate Mutase with Cofactor Homologues.
Angew.Chem.Int.Ed.Engl., 61, 2022
|
|
1XRS
 
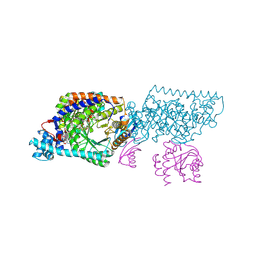 | | Crystal structure of Lysine 5,6-Aminomutase in complex with PLP, cobalamin, and 5'-deoxyadenosine | | Descriptor: | 5'-DEOXYADENOSINE, COBALAMIN, D-lysine 5,6-aminomutase alpha subunit, ... | | Authors: | Berkovitch, F, Behshad, E, Tang, K.H, Enns, E.A, Frey, P.A, Drennan, C.L. | | Deposit date: | 2004-10-15 | | Release date: | 2004-11-09 | | Last modified: | 2017-10-11 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | A locking mechanism preventing radical damage in the absence of substrate, as revealed by the x-ray structure of lysine 5,6-aminomutase.
Proc.Natl.Acad.Sci.Usa, 101, 2004
|
|
1BMT
 
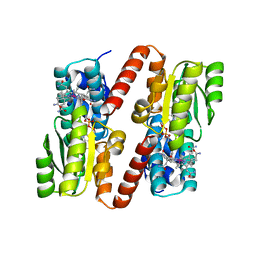 | | HOW A PROTEIN BINDS B12: A 3.O ANGSTROM X-RAY STRUCTURE OF THE B12-BINDING DOMAINS OF METHIONINE SYNTHASE | | Descriptor: | CO-METHYLCOBALAMIN, METHIONINE SYNTHASE | | Authors: | Drennan, C.L, Huang, S, Drummond, J.T, Matthews, R.G, Ludwig, M.L. | | Deposit date: | 1994-09-02 | | Release date: | 1995-06-03 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | How a protein binds B12: A 3.0 A X-ray structure of B12-binding domains of methionine synthase.
Science, 266, 1994
|
|
3KP1
 
 | | Crystal structure of ornithine 4,5 aminomutase (Resting State) | | Descriptor: | 5'-DEOXYADENOSINE, COBALAMIN, D-ornithine aminomutase E component, ... | | Authors: | Wolthers, K.R, Levy, C.W, Scrutton, N.S, Leys, D. | | Deposit date: | 2009-11-14 | | Release date: | 2010-01-26 | | Last modified: | 2019-10-30 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Large-scale domain dynamics and adenosylcobalamin reorientation orchestrate radical catalysis in ornithine 4,5-aminomutase.
J.Biol.Chem., 285, 2010
|
|
3KOW
 
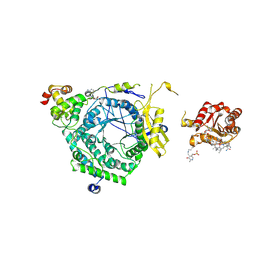 | | Crystal Structure of ornithine 4,5 aminomutase backsoaked complex | | Descriptor: | 5'-DEOXYADENOSINE, COBALAMIN, D-ornithine aminomutase E component, ... | | Authors: | Wolthers, K.R, Levy, C.W, Scrutton, N.S, Leys, D. | | Deposit date: | 2009-11-14 | | Release date: | 2010-01-26 | | Last modified: | 2012-10-24 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Large-scale domain dynamics and adenosylcobalamin reorientation orchestrate radical catalysis in ornithine 4,5-aminomutase.
J.Biol.Chem., 285, 2010
|
|
3KOX
 
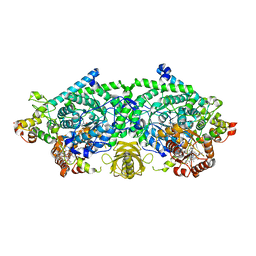 | | Crystal Structure of ornithine 4,5 aminomutase in complex with 2,4-diaminobutyrate (Anaerobic) | | Descriptor: | (2S)-2-amino-4-{[(1Z)-{3-hydroxy-2-methyl-5-[(phosphonooxy)methyl]pyridin-4-yl}methylidene]amino}butanoic acid, 5'-DEOXYADENOSINE, COBALAMIN, ... | | Authors: | Wolthers, K.R, Levy, C.W, Scrutton, N.S, Leys, D. | | Deposit date: | 2009-11-14 | | Release date: | 2010-01-26 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Large-scale domain dynamics and adenosylcobalamin reorientation orchestrate radical catalysis in ornithine 4,5-aminomutase.
J.Biol.Chem., 285, 2010
|
|
3KOZ
 
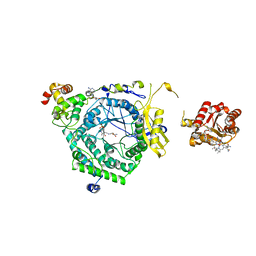 | | Crystal Structure of ornithine 4,5 aminomutase in complex with ornithine (Anaerobic) | | Descriptor: | (E)-N~5~-({3-hydroxy-2-methyl-5-[(phosphonooxy)methyl]pyridin-4-yl}methylidene)-L-ornithine, 5'-DEOXYADENOSINE, COBALAMIN, ... | | Authors: | Wolthers, K.R, Levy, C.W, Scrutton, N.S, Leys, D. | | Deposit date: | 2009-11-14 | | Release date: | 2010-01-26 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Large-scale domain dynamics and adenosylcobalamin reorientation orchestrate radical catalysis in ornithine 4,5-aminomutase.
J.Biol.Chem., 285, 2010
|
|
