4QUK
 
 | |
4R1U
 
 | | Crystal structure of Medicago truncatula cinnamoyl-CoA reductase | | Descriptor: | ACETATE ION, Cinnamoyl CoA reductase | | Authors: | Noel, J.P, Bomati, E.K, Louie, G.V, Bowman, M.E. | | Deposit date: | 2014-08-07 | | Release date: | 2014-10-01 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | Structural Studies of Cinnamoyl-CoA Reductase and Cinnamyl-Alcohol Dehydrogenase, Key Enzymes of Monolignol Biosynthesis.
Plant Cell, 26, 2014
|
|
4R1S
 
 | | Crystal structure of Petunia hydrida cinnamoyl-CoA reductase | | Descriptor: | NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, cinnamoyl CoA reductase | | Authors: | Noel, J.P, Louie, G.V, Bowman, M.E, Bomati, E.K. | | Deposit date: | 2014-08-07 | | Release date: | 2014-10-01 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structural Studies of Cinnamoyl-CoA Reductase and Cinnamyl-Alcohol Dehydrogenase, Key Enzymes of Monolignol Biosynthesis.
Plant Cell, 26, 2014
|
|
6WJA
 
 | |
6WJ9
 
 | | UDP-GlcNAc C4-epimerase mutant S121A/Y146F from Pseudomonas protegens in complex with UDP-GlcNAc | | Descriptor: | NAD-dependent epimerase/dehydratase family protein, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, URIDINE-DIPHOSPHATE-N-ACETYLGLUCOSAMINE | | Authors: | Marmont, L.S, Willams, R.J, Whitney, J.C, Whitfield, G.B, Robinson, H, Parsek, M.R, Nitz, M, Harrison, J.J, Howell, P.L. | | Deposit date: | 2020-04-13 | | Release date: | 2020-07-08 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | PelX is a UDP-N-acetylglucosamine C4-epimerase involved in Pel polysaccharide-dependent biofilm formation.
J.Biol.Chem., 295, 2020
|
|
6WJB
 
 | | UDP-GlcNAc C4-epimerase from Pseudomonas protegens in complex with NAD and UDP-GlcNAc | | Descriptor: | NAD-dependent epimerase/dehydratase family protein, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, URIDINE-DIPHOSPHATE-N-ACETYLGLUCOSAMINE | | Authors: | Marmont, L.S, Pfoh, R, Robinson, H, Howell, P.L. | | Deposit date: | 2020-04-13 | | Release date: | 2020-07-08 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | PelX is a UDP-N-acetylglucosamine C4-epimerase involved in Pel polysaccharide-dependent biofilm formation.
J.Biol.Chem., 295, 2020
|
|
6PNL
 
 | | Structure of Epimerase Mth375 from the thermophilic pseudomurein-containing methanogen Methanothermobacter thermautotrophicus | | Descriptor: | 1,2-ETHANEDIOL, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, SULFATE ION, ... | | Authors: | Carbone, V, Schofield, L.R, Sutherland-Smith, A.J, Ronimus, R.S. | | Deposit date: | 2019-07-02 | | Release date: | 2020-07-08 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | Structure of a UDP-GALE 4-epimerase (Mth375) from the thermophilic pseudomurein-containing methanogen Methanothermobacter thermautotrophicus
To Be Published
|
|
6PMH
 
 | | Structure of Epimerase Mth375 from the thermophilic pseudomurein-containing methanogen Methanothermobacter thermautotrophicus | | Descriptor: | ADENOSINE MONOPHOSPHATE, CHLORIDE ION, GLYCEROL, ... | | Authors: | Carbone, V, Schofield, L.R, Sutherland-Smith, A.J, Ronimus, R.S. | | Deposit date: | 2019-07-01 | | Release date: | 2020-07-08 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of a UDP-GALE 4-epimerase (Mth375) from the thermophilic pseudomurein-containing methanogen Methanothermobacter thermautotrophicus
To Be Published
|
|
1E7R
 
 | | GDP 4-keto-6-deoxy-D-mannose epimerase reductase Y136E | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ACETYLPHOSPHATE, GDP-FUCOSE SYNTHETASE, ... | | Authors: | Rosano, C, Izzo, G, Bolognesi, M. | | Deposit date: | 2000-09-07 | | Release date: | 2000-10-18 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Probing the Catalytic Mechanism of Gdp-4-Keto-6-Deoxy-D-Mannose Epimerase/Reductase by Kinetic and Crystallographic Characterization of Site-Specific Mutants
J.Mol.Biol., 303, 2000
|
|
1EQ2
 
 | | THE CRYSTAL STRUCTURE OF ADP-L-GLYCERO-D-MANNOHEPTOSE 6-EPIMERASE | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE-GLUCOSE, ADP-L-GLYCERO-D-MANNOHEPTOSE 6-EPIMERASE, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Deacon, A.M, Ni, Y.S, Coleman Jr, W.G, Ealick, S.E. | | Deposit date: | 2000-03-31 | | Release date: | 2000-11-08 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The crystal structure of ADP-L-glycero-D-mannoheptose 6-epimerase: catalysis with a twist.
Structure Fold.Des., 8, 2000
|
|
6JYG
 
 | | Crystal Structure of L-threonine dehydrogenase from Phytophthora infestans | | Descriptor: | 3,6,9,12,15,18,21-HEPTAOXATRICOSANE-1,23-DIOL, CITRATE ANION, L-threonine 3-dehydrogenase, ... | | Authors: | Yoneda, K, Sakuraba, H, Ohshima, T. | | Deposit date: | 2019-04-26 | | Release date: | 2020-04-01 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Catalytic properties and crystal structure of UDP-galactose 4-epimerase-like l-threonine 3-dehydrogenase from Phytophthora infestans.
Enzyme.Microb.Technol., 140, 2020
|
|
2C29
 
 | | Structure of dihydroflavonol reductase from Vitis vinifera at 1.8 A. | | Descriptor: | (2R,3R)-2-(3,4-DIHYDROXYPHENYL)-3,5,7-TRIHYDROXY-2,3-DIHYDRO-4H-CHROMEN-4-ONE, DIHYDROFLAVONOL 4-REDUCTASE, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Petit, P, Granier, T, D'Estaintot, B.L, Hamdi, S, Gallois, B. | | Deposit date: | 2005-09-27 | | Release date: | 2006-10-16 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Crystal Structure of Grape Dihydroflavonol 4-Reductase, a Key Enzyme in Flavonoid Biosynthesis.
J.Mol.Biol., 368, 2007
|
|
2BLL
 
 | | Apo-structure of the C-terminal decarboxylase domain of ArnA | | Descriptor: | PROTEIN YFBG | | Authors: | Williams, G.J, Breazeale, S.D, Raetz, C.R.H, Naismith, J.H. | | Deposit date: | 2005-03-07 | | Release date: | 2005-04-08 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure and Function of Both Domains of Arna, a Dual Function Decarboxylase and a Formyltransferase, Involved in 4-Amino-4-Deoxy-L- Arabinose Biosynthesis.
J.Biol.Chem., 280, 2005
|
|
2C59
 
 | | gdp-mannose-3', 5' -epimerase (arabidopsis thaliana), with gdp-alpha-d-mannose and gdp-beta-l-galactose bound in the active site. | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, FORMIC ACID, GDP-MANNOSE-3', ... | | Authors: | Major, L.L, Wolucka, B.A, Naismith, J.H. | | Deposit date: | 2005-10-26 | | Release date: | 2005-11-14 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure and Function of Gdp-Mannose-3',5'-Epimerase: An Enzyme which Performs Three Chemical Reactions at the Same Active Site.
J.Am.Chem.Soc., 127, 2005
|
|
6KWS
 
 | |
6KV9
 
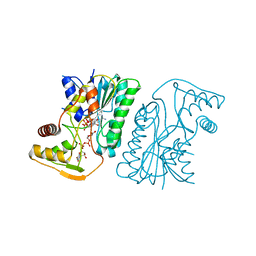 | | MoeE5 in complex with UDP-glucuronic acid and NAD | | Descriptor: | MoeE5, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, URIDINE-5'-DIPHOSPHATE-GLUCURONIC ACID | | Authors: | Ko, T.-P, Liu, W, Sun, H, Liu, W, Chen, C.-C, Guo, R.-T. | | Deposit date: | 2019-09-03 | | Release date: | 2019-11-13 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | Structure of an antibiotic-synthesizing UDP-glucuronate 4-epimerase MoeE5 in complex with substrate.
Biochem.Biophys.Res.Commun., 521, 2020
|
|
2C20
 
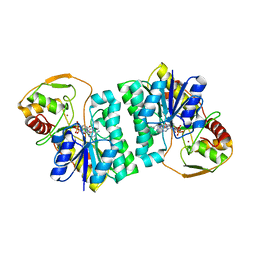 | | CRYSTAL STRUCTURE OF UDP-GLUCOSE 4-EPIMERASE | | Descriptor: | NICOTINAMIDE-ADENINE-DINUCLEOTIDE, UDP-GLUCOSE 4-EPIMERASE, ZINC ION | | Authors: | Lebedev, A.A, Moroz, O.V, Blagova, E.V, Levdikov, V.M, Fogg, M.J, Brannigan, J.A, Wilkinson, A.J, Wilson, K.S. | | Deposit date: | 2005-09-22 | | Release date: | 2007-02-20 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal Structure of Udp-Glucose 4-Epimerase from Bacillus Anthracis at 2.7A Resolution
To be Published
|
|
8H61
 
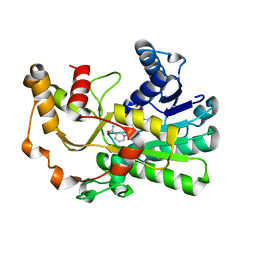 | | Ketoreductase CpKR mutant - M2 | | Descriptor: | Mutant M2 of ketoreductase CpKR, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Chen, C, Pan, J, Xu, J.H. | | Deposit date: | 2022-10-14 | | Release date: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Computational redesign of a robust ketoreductase for asymmetric synthesis of enantiopure diltiazem precursor.
To Be Published
|
|
2C5E
 
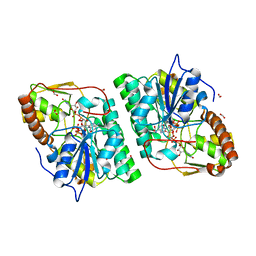 | | gdp-mannose-3', 5' -epimerase (arabidopsis thaliana), k217a, with gdp-alpha-d-mannose bound in the active site. | | Descriptor: | FORMIC ACID, GDP-MANNOSE-3', 5'-EPIMERASE, ... | | Authors: | Major, L.L, Wolucka, B.A, Naismith, J.H. | | Deposit date: | 2005-10-26 | | Release date: | 2005-11-14 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure and Function of Gdp-Mannose-3',5'-Epimerase: An Enzyme which Performs Three Chemical Reactions at the Same Active Site.
J.Am.Chem.Soc., 127, 2005
|
|
6KWT
 
 | |
2C5A
 
 | | GDP-mannose-3', 5' -epimerase (Arabidopsis thaliana),Y174F, with GDP-beta-L-galactose bound in the active site | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, FORMIC ACID, GDP-MANNOSE-3', ... | | Authors: | Major, L.L, Wolucka, B.A, Naismith, J.H. | | Deposit date: | 2005-10-26 | | Release date: | 2005-11-14 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structure and Function of Gdp-Mannose-3',5'-Epimerase: An Enzyme which Performs Three Chemical Reactions at the Same Active Site.
J.Am.Chem.Soc., 127, 2005
|
|
8GIL
 
 | | L-threonine 3-Dehydrogenase from Trypanosoma cruzi (apo form) | | Descriptor: | L-threonine 3-dehydrogenase, POTASSIUM ION | | Authors: | Faria, J.N, Mercaldi, G.F, Fagundes, M, Bezerra, E.H.S, Cordeiro, A.T. | | Deposit date: | 2023-03-14 | | Release date: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure, allosteric regulation and metabolic activity of L-threonine 3-Dehydrogenase from Trypanosoma cruzi
To Be Published
|
|
8GJB
 
 | | L-threonine 3-Dehydrogenase from Trypanosoma cruzi in complex with NAD and acetate | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ACETATE ION, GLYCEROL, ... | | Authors: | Mercaldi, G.F, Faria, J.N, Fagundes, M, Bezerra, E.H.S, Cordeiro, A.T. | | Deposit date: | 2023-03-15 | | Release date: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structure, allosteric regulation and metabolic activity of L-threonine 3-Dehydrogenase from Trypanosoma cruzi
To Be Published
|
|
2C54
 
 | | gdp-mannose-3', 5' -epimerase (arabidopsis thaliana),k178r, with gdp-beta-l-gulose and gdp-4-keto-beta-l-gulose bound in active site. | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, FORMIC ACID, GDP-MANNOSE-3', ... | | Authors: | Major, L.L, Wolucka, B.A, Naismith, J.H. | | Deposit date: | 2005-10-25 | | Release date: | 2005-11-14 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure and function of GDP-mannose-3',5'-epimerase: an enzyme which performs three chemical reactions at the same active site.
J. Am. Chem. Soc., 127, 2005
|
|
6KVC
 
 | | MoeE5 in complex with UDP-glucose and NAD | | Descriptor: | MoeE5, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, URIDINE-5'-DIPHOSPHATE-GLUCOSE | | Authors: | Ko, T.-P, Liu, W, Sun, H, Liu, W, Chen, C.-C, Guo, R.-T. | | Deposit date: | 2019-09-04 | | Release date: | 2019-11-13 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | Structure of an antibiotic-synthesizing UDP-glucuronate 4-epimerase MoeE5 in complex with substrate.
Biochem.Biophys.Res.Commun., 521, 2020
|
|
