6KBL
 
 | | Structure-function study of AKR4C14, an aldo-keto reductase from Thai Jasmine rice (Oryza sativa L. ssp. Indica cv. KDML105) | | 分子名称: | ACETATE ION, Aldo-keto reductase, CACODYLATE ION, ... | | 著者 | Songsiriritthigul, C, Narawongsanont, R, Guan, H.H, Chen, C.J. | | 登録日 | 2019-06-25 | | 公開日 | 2020-05-06 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Structure-function study of AKR4C14, an aldo-keto reductase from Thai jasmine rice (Oryza sativa L. ssp. indica cv. KDML105).
Acta Crystallogr D Struct Biol, 76, 2020
|
|
6OVQ
 
 | | Crystal structure of mithramycin 3-side chain keto-reductase MtmW | | 分子名称: | GLYCEROL, Putative Side chain reductase | | 著者 | Hou, C, Yu, X, Rohr, J, Tsodikov, O.V. | | 登録日 | 2019-05-08 | | 公開日 | 2019-11-27 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (1.8 Å) | | 主引用文献 | Discovery of a Cryptic Intermediate in Late Steps of Mithramycin Biosynthesis.
Angew.Chem.Int.Ed.Engl., 59, 2020
|
|
6OW0
 
 | | Crystal structure of mithramycin 3-side chain keto-reductase MtmW in complex with NAD+ and PEG | | 分子名称: | DI(HYDROXYETHYL)ETHER, GLYCEROL, MtmW, ... | | 著者 | Hou, C, Yu, X, Rohr, J, Tsodikov, O.V. | | 登録日 | 2019-05-08 | | 公開日 | 2019-11-27 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (2.67 Å) | | 主引用文献 | Discovery of a Cryptic Intermediate in Late Steps of Mithramycin Biosynthesis.
Angew.Chem.Int.Ed.Engl., 59, 2020
|
|
6OVX
 
 | | Crystal structure of mithramycin 3-side chain keto-reductase MtmW in complex with NAD+, P422 form | | 分子名称: | GLYCEROL, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, Putative side chain reductase | | 著者 | Hou, C, Yu, X, Rohr, J, Tsodikov, O.V. | | 登録日 | 2019-05-08 | | 公開日 | 2019-11-27 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Discovery of a Cryptic Intermediate in Late Steps of Mithramycin Biosynthesis.
Angew.Chem.Int.Ed.Engl., 59, 2020
|
|
6IJX
 
 | | Crystal Structure of AKR1C1 complexed with meclofenamic acid | | 分子名称: | 2-[(2,6-dichloro-3-methyl-phenyl)amino]benzoic acid, Aldo-keto reductase family 1 member C1, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | 著者 | Zheng, X, Zhao, Y, Zhang, L, Zhang, H, Chen, Y, Hu, X. | | 登録日 | 2018-10-12 | | 公開日 | 2019-10-16 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.2 Å) | | 主引用文献 | Screening, synthesis, crystal structure, and molecular basis of 6-amino-4-phenyl-1,4-dihydropyrano[2,3-c]pyrazole-5-carbonitriles as novel AKR1C3 inhibitors.
Bioorg.Med.Chem., 26, 2018
|
|
6KY6
 
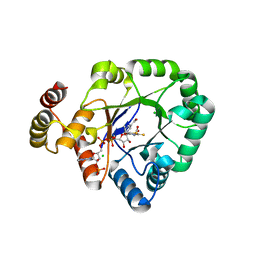 | | Crystal structure of a thermostable aldo-keto reductase Tm1743 in complexs with inhibitor epalrestat in space group P3221cc | | 分子名称: | 2,5-diketo-D-gluconic acid reductase, CHLORIDE ION, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | 著者 | Zhang, C.Y, Min, Z.Z, Liu, X.M, Wang, C, Tang, W.R. | | 登録日 | 2019-09-16 | | 公開日 | 2019-10-02 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.07 Å) | | 主引用文献 | Tolrestat acts atypically as a competitive inhibitor of the thermostable aldo-keto reductase Tm1743 from Thermotoga maritima.
Febs Lett., 594, 2020
|
|
6KIK
 
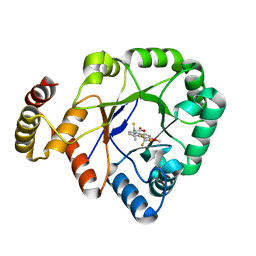 | | Crystal structure of a thermostable aldo-keto reductase Tm1743 in complex with inhibitor tolrestat | | 分子名称: | Oxidoreductase, aldo/keto reductase family, TOLRESTAT | | 著者 | Zhang, C.Y, Liu, X.M, Wang, C, Tang, W.R. | | 登録日 | 2019-07-18 | | 公開日 | 2019-09-25 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.601 Å) | | 主引用文献 | Tolrestat acts atypically as a competitive inhibitor of the thermostable aldo-keto reductase Tm1743 from Thermotoga maritima.
Febs Lett., 594, 2020
|
|
6KIY
 
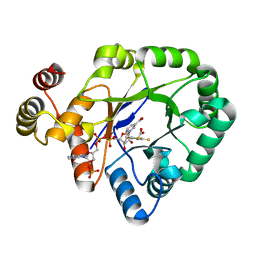 | | Crystal structure of a thermostable aldo-keto reductase Tm1743 in complex with inhibitor Epalrestat | | 分子名称: | NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, Oxidoreductase, aldo/keto reductase family, ... | | 著者 | Zhang, C.Y, Liu, X.M, Wang, C, Min, Z.Z, Xu, X.L. | | 登録日 | 2019-07-20 | | 公開日 | 2019-09-25 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (1.9 Å) | | 主引用文献 | Tolrestat acts atypically as a competitive inhibitor of the thermostable aldo-keto reductase Tm1743 from Thermotoga maritima.
Febs Lett., 594, 2020
|
|
6A7A
 
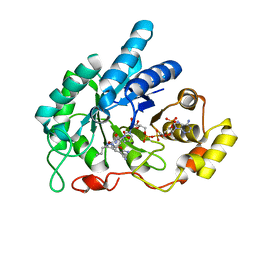 | | AKR1C1 complexed with new inhibitor with novel scaffold | | 分子名称: | (4R)-6-amino-4-(4-hydroxy-3-methoxy-5-nitrophenyl)-3-propyl-1,4-dihydropyrano[2,3-c]pyrazole-5-carbonitrile, Aldo-keto reductase family 1 member C1, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | 著者 | Zheng, X, Zhao, Y, Zhang, H, Chen, Y. | | 登録日 | 2018-07-02 | | 公開日 | 2019-07-03 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.37 Å) | | 主引用文献 | Screening, synthesis, crystal structure, and molecular basis of 6-amino-4-phenyl-1,4-dihydropyrano[2,3-c]pyrazole-5-carbonitriles as novel AKR1C3 inhibitors.
Bioorg.Med.Chem., 26, 2018
|
|
6A7B
 
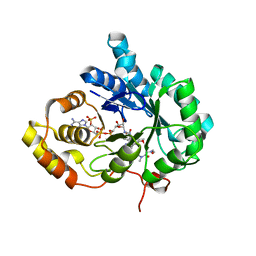 | | AKR1C3 complexed with new inhibitor with novel scaffold | | 分子名称: | (4R)-6-amino-4-(4-hydroxy-3-methoxy-5-nitrophenyl)-3-propyl-1,4-dihydropyrano[2,3-c]pyrazole-5-carbonitrile, Aldo-keto reductase family 1 member C3, DIMETHYLFORMAMIDE, ... | | 著者 | Zheng, X, Zhao, Y, Zhang, H, Chen, Y. | | 登録日 | 2018-07-02 | | 公開日 | 2019-07-03 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.37 Å) | | 主引用文献 | Screening, synthesis, crystal structure, and molecular basis of 6-amino-4-phenyl-1,4-dihydropyrano[2,3-c]pyrazole-5-carbonitriles as novel AKR1C3 inhibitors.
Bioorg.Med.Chem., 26, 2018
|
|
6HG6
 
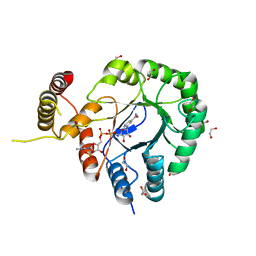 | | Clostridium beijerinckii aldo-keto reductase Cbei_3974 with NADPH | | 分子名称: | 1,2-ETHANEDIOL, L-glyceraldehyde 3-phosphate reductase, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | 著者 | Scott, A.F, Cresser-Brown, J, Rizkallah, P.J, Jin, Y, Allemann, R.K. | | 登録日 | 2018-08-22 | | 公開日 | 2019-05-29 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (1.75 Å) | | 主引用文献 | Crystal Structure and Biophysical Analysis of Furfural-Detoxifying Aldehyde Reductase from Clostridium beijerinckii.
Appl.Environ.Microbiol., 85, 2019
|
|
6GXK
 
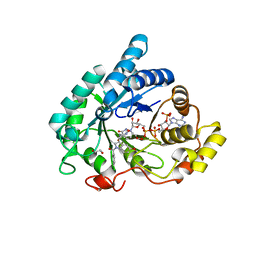 | | Crystal structure of Aldo-Keto Reductase 1C3 (AKR1C3) complexed with inhibitor. | | 分子名称: | 1,2-ETHANEDIOL, 4-[[1-(4-chlorophenyl)carbonyl-5-methoxy-2-methyl-indol-3-yl]methyl]-1,2,5-oxadiazol-3-one, Aldo-keto reductase family 1 member C3, ... | | 著者 | Goyal, P, Wahlgren, W.Y, Friemann, R. | | 登録日 | 2018-06-27 | | 公開日 | 2019-05-08 | | 最終更新日 | 2019-11-20 | | 実験手法 | X-RAY DIFFRACTION (1.7 Å) | | 主引用文献 | Bioisosteres of Indomethacin as Inhibitors of Aldo-Keto Reductase 1C3.
Acs Med.Chem.Lett., 10, 2019
|
|
6F84
 
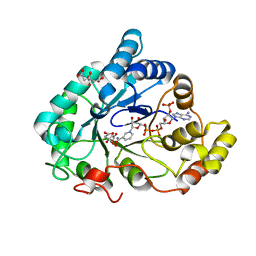 | | AKR1B1 at 2.55 MGy radiation dose. | | 分子名称: | Aldose reductase, CITRIC ACID, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | 著者 | Castellvi, A, Juanhuix, J. | | 登録日 | 2017-12-12 | | 公開日 | 2019-03-13 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (1.09 Å) | | 主引用文献 | Efficacy of aldose reductase inhibitors is affected by oxidative stress induced under X-ray irradiation.
Sci Rep, 9, 2019
|
|
6F8O
 
 | | AKR1B1 at 3.45 MGy radiation dose. | | 分子名称: | Aldose reductase, CITRIC ACID, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | 著者 | Castellvi, A, Juanhuix, J. | | 登録日 | 2017-12-13 | | 公開日 | 2019-03-13 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (1.17 Å) | | 主引用文献 | Efficacy of aldose reductase inhibitors is affected by oxidative stress induced under X-ray irradiation.
Sci Rep, 9, 2019
|
|
6F81
 
 | | AKR1B1 at 0.75 MGy radiation dose. | | 分子名称: | Aldose reductase, CITRIC ACID, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | 著者 | Castellvi, A, Juanhuix, J. | | 登録日 | 2017-12-12 | | 公開日 | 2019-03-13 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (0.97 Å) | | 主引用文献 | Efficacy of aldose reductase inhibitors is affected by oxidative stress induced under X-ray irradiation.
Sci Rep, 9, 2019
|
|
6F7R
 
 | | AKR1B1 at 0.03 MGy radiation dose. | | 分子名称: | Aldose reductase, CITRIC ACID, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | 著者 | Castellvi, A, Juanhuix, J. | | 登録日 | 2017-12-11 | | 公開日 | 2019-03-13 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (0.92 Å) | | 主引用文献 | Efficacy of aldose reductase inhibitors is affected by oxidative stress induced under X-ray irradiation.
Sci Rep, 9, 2019
|
|
6F82
 
 | | AKR1B1 at 1.65 MGy radiation dose. | | 分子名称: | Aldose reductase, CITRIC ACID, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | 著者 | Castellvi, A, Juanhuix, J. | | 登録日 | 2017-12-12 | | 公開日 | 2019-03-13 | | 最終更新日 | 2024-01-17 | | 実験手法 | X-RAY DIFFRACTION (1.03 Å) | | 主引用文献 | Efficacy of aldose reductase inhibitors is affected by oxidative stress induced under X-ray irradiation.
Sci Rep, 9, 2019
|
|
6CI1
 
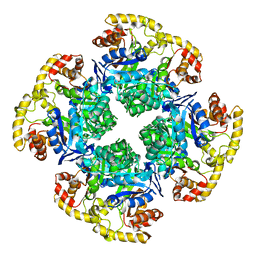 | |
6M7K
 
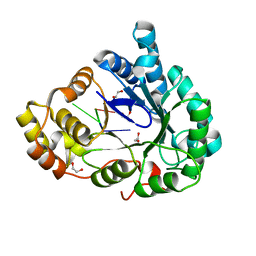 | | Structure of mouse RECON (AKR1C13) in complex with cyclic AMP-AMP-GMP (cAAG) | | 分子名称: | 1,2-ETHANEDIOL, Aldo-keto reductase family 1 member C13, cyclic AMP-AMP-GMP | | 著者 | Eaglesham, J.B, Whiteley, A.T, de Oliveira Mann, C.C, Morehouse, B.R, Nieminen, E.A, King, D.S, Lee, A.S.Y, Mekalanos, J.J, Kranzusch, P.J. | | 登録日 | 2018-08-20 | | 公開日 | 2019-02-20 | | 最終更新日 | 2023-10-11 | | 実験手法 | X-RAY DIFFRACTION (1.1 Å) | | 主引用文献 | Bacterial cGAS-like enzymes synthesize diverse nucleotide signals.
Nature, 567, 2019
|
|
5Z6T
 
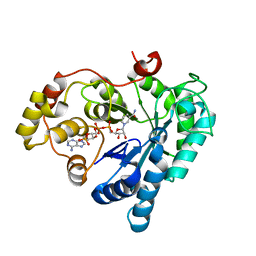 | |
5Z6U
 
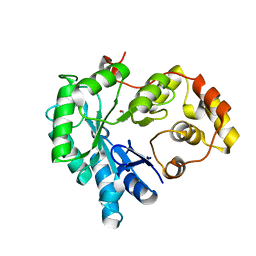 | |
5ZCI
 
 | |
5ZCM
 
 | |
5Y7N
 
 | | Crystal structure of AKR1B10 in complex with NADP+ and Androst-4-ene-3-beta-6-alpha-diol | | 分子名称: | (3S,6S,8S,9S,10R,13S,14R)-10,13-dimethyl-2,3,6,7,8,9,11,12,14,15,16,17-dodecahydro-1H-cyclopenta[a]phenanthrene-3,6-diol, Aldo-keto reductase family 1 member B10, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | 著者 | Hong, Z, Liping, Z, Cuiyun, L, Wei, Z. | | 登録日 | 2017-08-17 | | 公開日 | 2018-08-29 | | 最終更新日 | 2023-11-22 | | 実験手法 | X-RAY DIFFRACTION (2.5 Å) | | 主引用文献 | Structural determinants for the inhibition of AKR1B10 by steroids
To Be Published
|
|
6EBK
 
 | | The voltage-activated Kv1.2-2.1 paddle chimera channel in lipid nanodiscs | | 分子名称: | NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, Potassium voltage-gated channel subfamily A member 2,Potassium voltage-gated channel subfamily B member 2 chimera, Voltage-gated potassium channel subunit beta-2 | | 著者 | Matthies, D, Bae, C, Fox, T, Bartesaghi, A, Subramaniam, S, Swartz, K.J. | | 登録日 | 2018-08-06 | | 公開日 | 2018-08-22 | | 最終更新日 | 2024-03-13 | | 実験手法 | ELECTRON MICROSCOPY (3.3 Å) | | 主引用文献 | Single-particle cryo-EM structure of a voltage-activated potassium channel in lipid nanodiscs.
Elife, 7, 2018
|
|
