5OU0
 
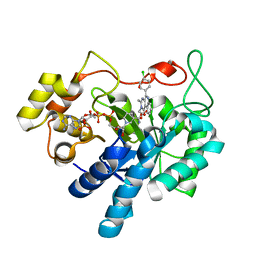 | | Crystal structure of human AKR1B1 complexed with NADP+ and compound 37 | | Descriptor: | 2-[5-(4-chlorophenyl)-3-methyl-1-oxidanylidene-pyrimido[4,5-c]quinolin-2-yl]ethanoic acid, Aldose reductase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Cousido-Siah, A, Ruiz, F.X, Mitschler, A, Metwally, K, Podjarny, A. | | Deposit date: | 2017-08-23 | | Release date: | 2018-05-09 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (0.94 Å) | | Cite: | Design, synthesis, structure-activity relationships and X-ray structural studies of novel 1-oxopyrimido[4,5-c]quinoline-2-acetic acid derivatives as selective and potent inhibitors of human aldose reductase.
Eur J Med Chem, 152, 2018
|
|
4MHB
 
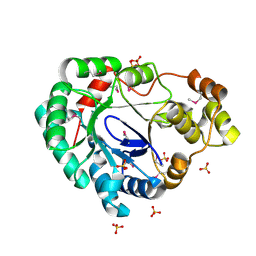 | | Structure of a putative reductase from Yersinia pestis | | Descriptor: | Putative aldo/keto reductase, SULFATE ION | | Authors: | Anderson, S.M, Wawrzak, Z, Kudritska, M, Kwon, K, Rembert, P, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2013-08-29 | | Release date: | 2013-10-16 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structure of a putative reductase from Yersinia pestis
To be Published
|
|
1GVE
 
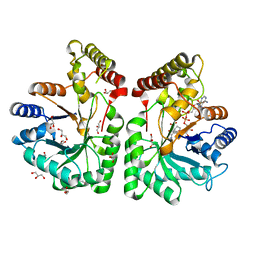 | | Aflatoxin aldehyde reductase (AKR7A1) from Rat Liver | | Descriptor: | AFLATOXIN B1 ALDEHYDE REDUCTASE MEMBER 3, CITRIC ACID, GLYCEROL, ... | | Authors: | Kozma, E, Brown, E, Ellis, E.M, Lapthorn, A.J. | | Deposit date: | 2002-02-08 | | Release date: | 2002-06-27 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.38 Å) | | Cite: | The Crystal Structure of Rat Liver Akr7A1: A Dimeric Member of the Aldo-Keto Reductase Superfamily
J.Biol.Chem., 277, 2002
|
|
1QRQ
 
 | |
8RB6
 
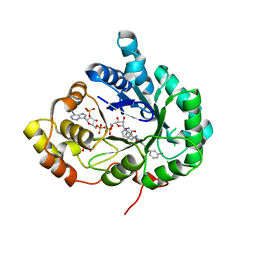 | | Structure of Aldo-Keto Reductase 1C3 (AKR1C3) in complex with an inhibitor M689, with the 3-hydroxy-benzoisoxazole moiety. Resolution 2.0A | | Descriptor: | 1,2-ETHANEDIOL, 4-[[4-(3-hydroxyphenyl)phenyl]amino]-1,2-benzoxazol-3-ol, Aldo-keto reductase family 1 member C3, ... | | Authors: | Frydenvang, K, Mirza, O.A. | | Deposit date: | 2023-12-03 | | Release date: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure-guided optimization of 3-hydroxybenzoisoxazole derivatives as inhibitors of Aldo-keto reductase 1C3 (AKR1C3) to target prostate cancer.
Eur.J.Med.Chem., 268, 2024
|
|
6XUM
 
 | | Human Aldose Reductase Mutant L300/301A in Complex with a Ligand with an IDD Structure ({5-fluoro-2-[(3-nitrobenzyl)carbamoyl]phenoxy}acetic acid) | | Descriptor: | Aldo-keto reductase family 1 member B1, CITRIC ACID, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Hubert, L.-S, Ley, M, Heine, A, Klebe, G. | | Deposit date: | 2020-01-20 | | Release date: | 2021-02-03 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (0.97 Å) | | Cite: | Human Aldose Reductase Mutant L300/301A in Complex with a Ligand with an IDD Structure ({5-fluoro-2-[(3-nitrobenzyl)carbamoyl]phenoxy}acetic acid)
To Be Published
|
|
5T79
 
 | | X-Ray Crystal Structure of a Novel Aldo-keto Reductases for the Biocatalytic Conversion of 3-hydroxybutanal to 1,3-butanediol | | Descriptor: | Aldo-keto Reductase, OXIDOREDUCTASE, CHLORIDE ION, ... | | Authors: | Brunzelle, J.S, Wawrzak, Z, Evdokimova, E, Kudritska, M, Savchenko, A, Yakunin, A.F, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2016-09-02 | | Release date: | 2017-02-15 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Structural and biochemical studies of novel aldo-keto reductases for the biocatalytic conversion of 3-hydroxybutanal to 1,3-butanediol.
Appl. Environ. Microbiol., 2017
|
|
3BUV
 
 | | Crystal structure of human Delta(4)-3-ketosteroid 5-beta-reductase in complex with NADP and HEPES. Resolution: 1.35 A. | | Descriptor: | 3-oxo-5-beta-steroid 4-dehydrogenase, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, GLYCEROL, ... | | Authors: | Di Costanzo, L, Drury, J, Penning, T.M, Christianson, D.W. | | Deposit date: | 2008-01-03 | | Release date: | 2008-04-01 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Crystal Structure of Human Liver {Delta}4-3-Ketosteroid 5{beta}-Reductase (AKR1D1) and Implications for Substrate Binding and Catalysis.
J.Biol.Chem., 283, 2008
|
|
3C3U
 
 | | Crystal structure of AKR1C1 in complex with NADP and 3,5-dichlorosalicylic acid | | Descriptor: | 3,5-dichloro-2-hydroxybenzoic acid, Aldo-keto reductase family 1 member C1, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Dhagat, U, El-Kabbani, O. | | Deposit date: | 2008-01-28 | | Release date: | 2008-08-26 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Selectivity determinants of inhibitor binding to human 20alpha-hydroxysteroid dehydrogenase: crystal structure of the enzyme in ternary complex with coenzyme and the potent inhibitor 3,5-dichlorosalicylic acid
J.Med.Chem., 51, 2008
|
|
5C7H
 
 | | Crystal structure of aldo-keto reductase from Sinorhizobium meliloti 1021 in complex with NADPH | | Descriptor: | Aldo-keto reductase, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Gasiorowska, O.A, Shabalin, I.G, Handing, K.B, Seidel, R, Bonanno, J, Almo, S.C, Minor, W, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2015-06-24 | | Release date: | 2015-07-08 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Crystal structure of aldo-keto reductase from Sinorhizobium meliloti 1021 in complex with NADPH
to be published
|
|
4WDU
 
 | | 17beta-HSD5 in complex with 4-chloro-N-(4-chlorobenzyl)-5-nitro-1H-pyrazole-3-carboxamide | | Descriptor: | 4-chloro-N-(4-chlorobenzyl)-5-nitro-1H-pyrazole-3-carboxamide, Aldo-keto reductase family 1 member C3, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Amano, Y, Yamaguchi, T. | | Deposit date: | 2014-09-09 | | Release date: | 2015-04-15 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structures of complexes of type 5 17 beta-hydroxysteroid dehydrogenase with structurally diverse inhibitors: insights into the conformational changes upon inhibitor binding.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
5JH2
 
 | | Crystal structure of the holo form of AKR4C7 from maize | | Descriptor: | 1,2-ETHANEDIOL, ADENOSINE-2'-5'-DIPHOSPHATE, Aldose reductase, ... | | Authors: | Giuseppe, P.O, Santos, M.L, Sousa, S.M, Koch, K.E, Yunes, J.A, Aparicio, R, Murakami, M.T. | | Deposit date: | 2016-04-20 | | Release date: | 2016-11-09 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | A comparative structural analysis reveals distinctive features of co-factor binding and substrate specificity in plant aldo-keto reductases.
Biochem.Biophys.Res.Commun., 474, 2016
|
|
4WDX
 
 | | 17beta-HSD5 in complex with [4-(2-hydroxyethyl)piperidin-1-yl](5-methyl-1H-indol-2-yl)methanone | | Descriptor: | Aldo-keto reductase family 1 member C3, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, [4-(2-hydroxyethyl)piperidin-1-yl](5-methyl-1H-indol-2-yl)methanone | | Authors: | Amano, Y, Yamaguchi, T. | | Deposit date: | 2014-09-09 | | Release date: | 2015-04-15 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Structures of complexes of type 5 17 beta-hydroxysteroid dehydrogenase with structurally diverse inhibitors: insights into the conformational changes upon inhibitor binding.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
8CNP
 
 | |
5JGY
 
 | | Crystal structure of maize AKR4C13 in P21 space group | | Descriptor: | 1,2-ETHANEDIOL, 4-{[(2R,3R,4S,5R)-5-({[(R)-{[(R)-{[(2R,3R,4R,5R)-5-(6-amino-9H-purin-9-yl)-3-hydroxy-4-(phosphonooxy)tetrahydrofuran-2-yl]methoxy}(hydroxy)phosphoryl]oxy}(hydroxy)phosphoryl]oxy}methyl)-3,4-dihydroxytetrahydrofuran-2-yl]oxy}butanoic acid (non-preferred name), Aldose reductase, ... | | Authors: | Santos, M.L, Giuseppe, P.O, Kiyota, E, Sousa, S.M, Schmelz, E.A, Yunes, J.A, Koch, K.E, Murakami, M.T, Aparicio, R. | | Deposit date: | 2016-04-20 | | Release date: | 2017-05-03 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Crystal structure of maize AKR4C13 in P21 space group
To Be Published
|
|
8CNF
 
 | |
6Y03
 
 | |
8OZ2
 
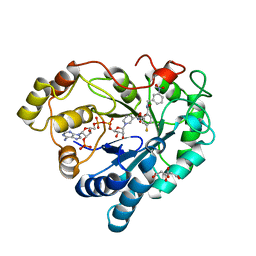 | |
6Y1P
 
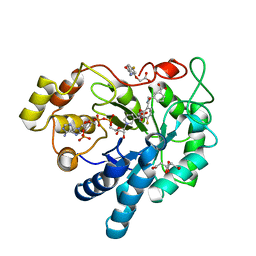 | | Human Aldose Reductase Mutant L300/301A in Complex with a Ligand with an IDD Structure (3-({[2-(carboxymethoxy)-4-fluorobenzoyl]amino}methyl)benzoic acid) | | Descriptor: | 3-({[2-(carboxymethoxy)-4-fluorobenzoyl]amino}methyl)benzoic acid, Aldo-keto reductase family 1 member B1, CITRIC ACID, ... | | Authors: | Hubert, L.-S, Ley, M, Heine, A, Klebe, G. | | Deposit date: | 2020-02-13 | | Release date: | 2021-02-24 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (0.94 Å) | | Cite: | Human Aldose Reductase Mutant L300/301A in Complex with a Ligand with an IDD Structure (3-({[2-(carboxymethoxy)-4-fluorobenzoyl]amino}methyl)benzoic acid)
To Be Published
|
|
8P2R
 
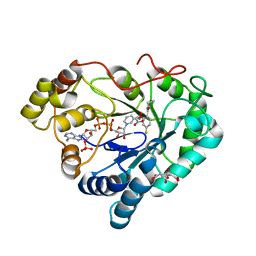 | |
5UXF
 
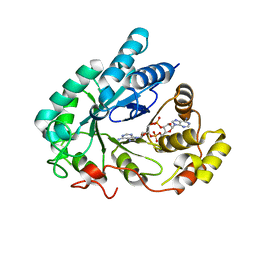 | | Crystal Structure of mouse RECON (AKR1C13) in complex with Cyclic di-AMP | | Descriptor: | (2R,3R,3aS,5R,7aR,9R,10R,10aS,12R,14aR)-2,9-bis(6-amino-9H-purin-9-yl)octahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8 ]tetraoxadiphosphacyclododecine-3,5,10,12-tetrol 5,12-dioxide, Dihydrodiol dehydrogenase | | Authors: | Luo, S, Tong, L. | | Deposit date: | 2017-02-22 | | Release date: | 2017-04-19 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.501 Å) | | Cite: | Sensing of Bacterial Cyclic Dinucleotides by the Oxidoreductase RECON Promotes NF-kappa B Activation and Shapes a Proinflammatory Antibacterial State.
Immunity, 46, 2017
|
|
4AUB
 
 | |
7LH6
 
 | |
4AST
 
 | |
7MBF
 
 | | codeinone reductase isoform 1.3 Apo form | | Descriptor: | NADPH-dependent codeinone reductase 1-3 | | Authors: | Carr, S.C, Ng, K.K.S. | | Deposit date: | 2021-03-31 | | Release date: | 2021-09-29 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural studies of codeinone reductase reveal novel insights into aldo-keto reductase function in benzylisoquinoline alkaloid biosynthesis.
J.Biol.Chem., 297, 2021
|
|
