5GZW
 
 | | Crystal structure of AmpC BER adenylylated by acetyl-AMP | | Descriptor: | ADENOSINE MONOPHOSPHATE, Beta-lactamase, SULFATE ION | | Authors: | An, Y.J, Cha, S.S. | | Deposit date: | 2016-10-02 | | Release date: | 2017-10-11 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.489 Å) | | Cite: | Structural and mechanistic insights into the inhibition of class C beta-lactamases through the adenylylation of the nucleophilic serine.
J.Antimicrob.Chemother., 72, 2017
|
|
5GSC
 
 | |
5GMX
 
 | | Crystal structure of a family VIII carboxylesterase | | Descriptor: | Carboxylesterase, phenylmethanesulfonic acid | | Authors: | An, Y.J, Cha, S.S. | | Deposit date: | 2016-07-18 | | Release date: | 2017-07-26 | | Last modified: | 2019-02-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of EstSRT1, a family VIII carboxylesterase displaying hydrolytic activity toward oxyimino cephalosporins
Biochem. Biophys. Res. Commun., 478, 2016
|
|
5GLD
 
 | | Crystal structure of the class A beta-lactamase PenL-tTR11 in complex with CBA | | Descriptor: | Beta-lactamase, PINACOL[[2-AMINO-ALPHA-(1-CARBOXY-1-METHYLETHOXYIMINO)-4-THIAZOLEACETYL]AMINO]METHANEBORONATE | | Authors: | Choi, J.M, Yi, H, Kim, H.S, Lee, S.H. | | Deposit date: | 2016-07-10 | | Release date: | 2017-02-15 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | High adaptability of the omega loop underlies the substrate-spectrum-extension evolution of a class A beta-lactamase, PenL
Sci Rep, 6, 2016
|
|
5GLC
 
 | | Crystal structure of the class A beta-lactamase PenL-tTR11 containing 20 residues insertion in omega-loop | | Descriptor: | Beta-lactamase | | Authors: | Choi, J.M, Yi, H, Kim, H.S, Lee, S.H. | | Deposit date: | 2016-07-10 | | Release date: | 2017-02-15 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.601 Å) | | Cite: | High adaptability of the omega loop underlies the substrate-spectrum-extension evolution of a class A beta-lactamase, PenL
Sci Rep, 6, 2016
|
|
5GLB
 
 | | Crystal structure of the class A beta-lactamase PenL-tTR10 in complex with CBA | | Descriptor: | Beta-lactamase, PINACOL[[2-AMINO-ALPHA-(1-CARBOXY-1-METHYLETHOXYIMINO)-4-THIAZOLEACETYL]AMINO]METHANEBORONATE | | Authors: | Choi, J.M, Yi, H, Kim, H.S, Lee, S.H. | | Deposit date: | 2016-07-10 | | Release date: | 2017-02-15 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | High adaptability of the omega loop underlies the substrate-spectrum-extension evolution of a class A beta-lactamase, PenL
Sci Rep, 6, 2016
|
|
5GLA
 
 | | Crystal structure of the class A beta-lactamase PenL-tTR10 containing 10 residues insertion in omega-loop | | Descriptor: | Beta-lactamase | | Authors: | Choi, J.M, Yi, H, Kim, H.S, Lee, S.H. | | Deposit date: | 2016-07-10 | | Release date: | 2017-02-15 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | High adaptability of the omega loop underlies the substrate-spectrum-extension evolution of a class A beta-lactamase, PenL
Sci Rep, 6, 2016
|
|
5GL9
 
 | | Crystal structure of the class A beta-lactamase PenL | | Descriptor: | Beta-lactamase, GLYCEROL | | Authors: | Choi, J.M, Yi, H, Kim, H.S, Lee, S.H. | | Deposit date: | 2016-07-10 | | Release date: | 2017-02-15 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | High adaptability of the omega loop underlies the substrate-spectrum-extension evolution of a class A beta-lactamase, PenL
Sci Rep, 6, 2016
|
|
5GKV
 
 | | Crystal Structure of a Novel Penicillin-Binding Protein (PBP) Homolog from Caulobacter crescentus | | Descriptor: | Esterase A | | Authors: | Ngo, T.D, Ryu, B.H, Kim, B.Y, Yoo, W.K, Lee, E.J, Lee, S.J, Kim, T.D, Kim, K.K. | | Deposit date: | 2016-07-07 | | Release date: | 2017-07-12 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.901 Å) | | Cite: | Biochemical and Structural Analysis of a Novel Penicillin-Binding Protein (PBP) Homolog from Caulobacter crescentus
To Be Published
|
|
5GHZ
 
 | |
5GHY
 
 | |
5GHX
 
 | | Crystal structure of beta-lactamase PenP mutant-E166H | | Descriptor: | 1,2-ETHANEDIOL, ACETIC ACID, Beta-lactamase | | Authors: | Pan, X, Zhao, Y. | | Deposit date: | 2016-06-21 | | Release date: | 2017-01-25 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.24 Å) | | Cite: | Crystallographic Snapshots of Class A beta-Lactamase Catalysis Reveal Structural Changes That Facilitate beta-Lactam Hydrolysis
J. Biol. Chem., 292, 2017
|
|
5GGW
 
 | | Crystal structure of Class C beta-lactamase | | Descriptor: | Beta-lactamase, PHOSPHATE ION, SULFATE ION | | Authors: | An, Y.J, Na, J.H, Cha, S.S. | | Deposit date: | 2016-06-16 | | Release date: | 2017-05-03 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.762 Å) | | Cite: | Structural basis for the extended substrate spectrum of AmpC BER and structure-guided discovery of the inhibition activity of citrate against the class C beta-lactamases AmpC BER and CMY-10.
Acta Crystallogr D Struct Biol, 72, 2016
|
|
5G18
 
 | | Direct Observation of Active-site Protonation States in a Class A beta lactamase with a monobactam substrate | | Descriptor: | 2-({[(1Z)-1-(2-amino-1,3-thiazol-4-yl)-2-oxo-2-{[(2S,3S)-1-oxo-3-(sulfoamino)butan-2-yl]amino}ethylidene]amino}oxy)-2-methylpropanoic acid, BETA-LACTAMASE CTX-M-97, SULFATE ION | | Authors: | Vandavasi, V.G, Weiss, K.L, Parks, J.M, Cooper, J.B, Ginell, S.L, Coates, L. | | Deposit date: | 2016-03-23 | | Release date: | 2016-11-09 | | Last modified: | 2019-10-23 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Active-Site Protonation States in an Acyl-Enzyme Intermediate of a Class A beta-Lactamase with a Monobactam Substrate.
Antimicrob. Agents Chemother., 61, 2017
|
|
5FAP
 
 | | CTX-M-15 in complex with FPI-1602 | | Descriptor: | Beta-lactamase, [[(3~{R},6~{S})-6-[(azetidin-3-ylcarbonylamino)carbamoyl]-1-methanoyl-piperidin-3-yl]amino] hydrogen sulfate | | Authors: | King, A.M, King, D.T, French, S, Brouillette, E, Asli, A, Alexander, A.N, Vuckovic, M, Maiti, S.N, Parr, T.R, Brown, E.D, Malouin, F, Strynadka, N.C.J, Wright, G.D. | | Deposit date: | 2015-12-11 | | Release date: | 2016-01-20 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural and Kinetic Characterization of Diazabicyclooctanes as Dual Inhibitors of Both Serine-beta-Lactamases and Penicillin-Binding Proteins.
Acs Chem.Biol., 11, 2016
|
|
5FAO
 
 | | CTX-M-15 in complex with FPI-1465 | | Descriptor: | Beta-lactamase, [[(3~{R},6~{S})-1-methanoyl-6-[[(3~{S})-pyrrolidin-3-yl]oxycarbamoyl]piperidin-3-yl]amino] hydrogen sulfate | | Authors: | King, A.M, King, D.T, French, S, Brouillette, E, Asli, A, Alexander, A.N, Vuckovic, M, Maiti, S.N, Parr, T.R, Brown, E.D, Malouin, F, Strynadka, N.C.J, Wright, G.D. | | Deposit date: | 2015-12-11 | | Release date: | 2016-02-17 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.01 Å) | | Cite: | Structural and Kinetic Characterization of Diazabicyclooctanes as Dual Inhibitors of Both Serine-beta-Lactamases and Penicillin-Binding Proteins.
Acs Chem.Biol., 11, 2016
|
|
5FA7
 
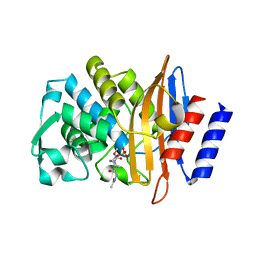 | | CTX-M-15 in complex with FPI-1523 | | Descriptor: | Beta-lactamase, [[(3~{R},6~{S})-6-(acetamidocarbamoyl)-1-methanoyl-piperidin-3-yl]amino] hydrogen sulfate | | Authors: | King, A.M, King, D.T, French, S, Brouillette, E, Asli, A, Alexander, A.N, Vuckovic, M, Maiti, S.N, Parr, T.R, Brown, E.D, Malouin, F, Strynadka, N.C.J, Wright, G.D. | | Deposit date: | 2015-12-11 | | Release date: | 2016-01-20 | | Last modified: | 2019-11-20 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Structural and Kinetic Characterization of Diazabicyclooctanes as Dual Inhibitors of Both Serine-beta-Lactamases and Penicillin-Binding Proteins.
Acs Chem.Biol., 11, 2016
|
|
5F83
 
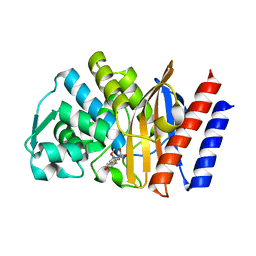 | | Imipenem complex of the GES-5 C69G mutant | | Descriptor: | (5R)-5-[(1S,2R)-1-formyl-2-hydroxypropyl]-3-[(2-{[(E)-iminomethyl]amino}ethyl)sulfanyl]-4,5-dihydro-1H-pyrrole-2-carboxylic acid, Beta-lactamase | | Authors: | Smith, C.A, Vakulenko, S.B. | | Deposit date: | 2015-12-09 | | Release date: | 2016-09-07 | | Last modified: | 2019-12-11 | | Method: | X-RAY DIFFRACTION (1.38 Å) | | Cite: | Role of the Conserved Disulfide Bridge in Class A Carbapenemases.
J.Biol.Chem., 291, 2016
|
|
5F82
 
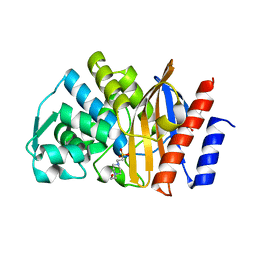 | | Apo GES-5 C69G mutant | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, Beta-lactamase | | Authors: | Smith, C.A, Vakulenko, S.B. | | Deposit date: | 2015-12-08 | | Release date: | 2016-09-07 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (0.96 Å) | | Cite: | Role of the Conserved Disulfide Bridge in Class A Carbapenemases.
J.Biol.Chem., 291, 2016
|
|
5F1G
 
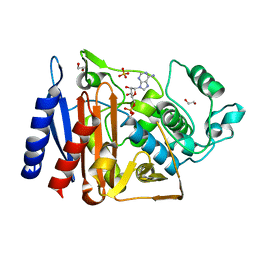 | | Crystal structure of AmpC BER adenylylated in the cytoplasm | | Descriptor: | 1,2-ETHANEDIOL, ADENOSINE MONOPHOSPHATE, Beta-lactamase, ... | | Authors: | An, Y.J, Kim, M.K, Na, J.H, Cha, S.S. | | Deposit date: | 2015-11-30 | | Release date: | 2016-12-07 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Structural and mechanistic insights into the inhibition of class C beta-lactamases through the adenylylation of the nucleophilic serine.
J.Antimicrob.Chemother., 72, 2017
|
|
5F1F
 
 | | Crystal structure of CMY-10 adenylylated by acetyl-AMP | | Descriptor: | ADENOSINE MONOPHOSPHATE, Beta-lactamase, CADMIUM ION | | Authors: | An, Y.J, Kim, M.K, Na, J.H, Cha, S.S. | | Deposit date: | 2015-11-30 | | Release date: | 2016-12-07 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.548 Å) | | Cite: | Structural and mechanistic insights into the inhibition of class C beta-lactamases through the adenylylation of the nucleophilic serine.
J.Antimicrob.Chemother., 72, 2017
|
|
5EVL
 
 | | Crystal Structure of Beta-Lactamase/D-Alanine Carboxypeptidase from Chromobacterium violaceum | | Descriptor: | Beta-lactamase, CHLORIDE ION, GLYCEROL, ... | | Authors: | Kim, Y, Hatzos-Skintges, C, Endres, M, Babnigg, G, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2015-11-19 | | Release date: | 2015-12-02 | | Last modified: | 2019-11-27 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | Crystal Structure of Beta-Lactamase/D-Alanine Carboxypeptidase from Chromobacterium violaceum
To Be Published
|
|
5EVI
 
 | | Crystal Structure of Beta-Lactamase/D-Alanine Carboxypeptidase from Pseudomonas syringae | | Descriptor: | 1,2-ETHANEDIOL, Beta-Lactamase/D-Alanine Carboxypeptidase, SULFATE ION | | Authors: | Kim, Y, Hatzos-Skintges, C, Endres, M, Babnigg, G, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2015-11-19 | | Release date: | 2016-01-13 | | Last modified: | 2019-12-04 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structure of Beta-Lactamase/D-Alanine Carboxypeptidase from Pseudomonas syringae
To Be Published
|
|
5EUA
 
 | | Crystal structure of extended-spectrum beta-lactamase BEL-1 in complex with Moxalactam | | Descriptor: | (2R)-2-[(1R)-1-{[(2R)-2-carboxy-2-(4-hydroxyphenyl)acetyl]amino}-1-methoxy-2-oxoethyl]-5-methylidene-5,6-dihydro-2H-1,3 -oxazine-4-carboxylic acid, Beta-lactamase, SODIUM ION | | Authors: | Pozzi, C, De Luca, F, Benvenuti, M, Docquier, J.D, Mangani, S. | | Deposit date: | 2015-11-18 | | Release date: | 2016-09-21 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal Structure of the Pseudomonas aeruginosa BEL-1 Extended-Spectrum beta-Lactamase and Its Complexes with Moxalactam and Imipenem.
Antimicrob.Agents Chemother., 60, 2016
|
|
5EPH
 
 | | Crystal structure of extended-spectrum beta-lactamase BEL-1 in complex with Imipenem | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, Beta-lactamase, CHLORIDE ION, ... | | Authors: | Pozzi, C, Benvenuti, M, De Luca, F, Docquier, J.D, Mangani, S. | | Deposit date: | 2015-11-11 | | Release date: | 2016-09-21 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Crystal Structure of the Pseudomonas aeruginosa BEL-1 Extended-Spectrum beta-Lactamase and Its Complexes with Moxalactam and Imipenem.
Antimicrob.Agents Chemother., 60, 2016
|
|
