7EOQ
 
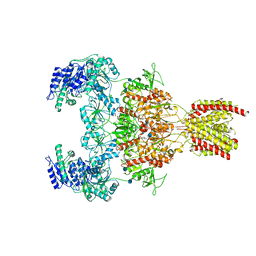 | |
7EOR
 
 | | Structure of the human GluN1/GluN2A NMDA receptor in the glycine/glutamate/GNE-6901 bound state | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 7-[(4-fluoranylphenoxy)methyl]-3-[(1~{R},2~{R})-2-(hydroxymethyl)cyclopropyl]-2-methyl-[1,3]thiazolo[3,2-a]pyrimidin-5-one, Glutamate receptor ionotropic, ... | | Authors: | Wang, H, Zhu, S. | | Deposit date: | 2021-04-22 | | Release date: | 2021-06-30 | | Last modified: | 2021-08-18 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Gating mechanism and a modulatory niche of human GluN1-GluN2A NMDA receptors.
Neuron, 109, 2021
|
|
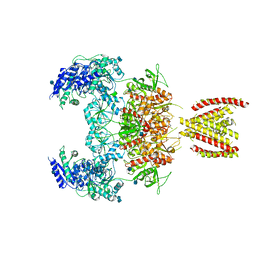
7EU8
 
 | | Structure of the human GluN1-GluN2B NMDA receptor in complex with S-ketamine,glycine and glutamate | | Descriptor: | (2~{S})-2-(2-chlorophenyl)-2-(methylamino)cyclohexan-1-one, 2-acetamido-2-deoxy-beta-D-glucopyranose, Glutamate receptor ionotropic, ... | | Authors: | Zhang, T, Zhang, Y, Zhu, S. | | Deposit date: | 2021-05-16 | | Release date: | 2021-07-28 | | Last modified: | 2022-10-26 | | Method: | ELECTRON MICROSCOPY (4.07 Å) | | Cite: | Structural basis of ketamine action on human NMDA receptors.
Nature, 596, 2021
|
|
7F3O
 
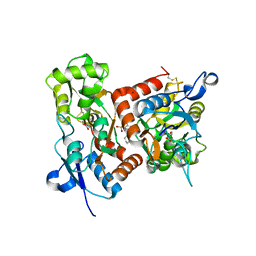 | | Crystal structure of the GluA2o LBD in complex with glutamate and TAK-653 | | Descriptor: | 7-(4-cyclohexyloxyphenyl)-9-methyl-4$l^{6}-thia-1$l^{4},5,8-triazabicyclo[4.4.0]deca-1(10),6,8-triene 4,4-dioxide, ACETATE ION, GLUTAMIC ACID, ... | | Authors: | Sogabe, S, Igaki, S, Hirokawa, A, Zama, Y, Lane, W, Snell, G. | | Deposit date: | 2021-06-16 | | Release date: | 2021-07-28 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.44 Å) | | Cite: | Strictly regulated agonist-dependent activation of AMPA-R is the key characteristic of TAK-653 for robust synaptic responses and cognitive improvement.
Sci Rep, 11, 2021
|
|
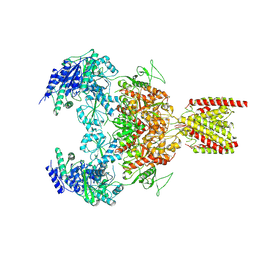
7EU7
 
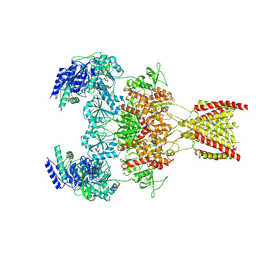 | | Structure of the human GluN1-GluN2A NMDA receptor in complex with S-ketamine, glycine and glutamate | | Descriptor: | (2~{S})-2-(2-chlorophenyl)-2-(methylamino)cyclohexan-1-one, 2-acetamido-2-deoxy-beta-D-glucopyranose, GLUTAMIC ACID, ... | | Authors: | Zhang, Y, Zhang, T, Zhu, S. | | Deposit date: | 2021-05-16 | | Release date: | 2021-08-04 | | Last modified: | 2022-10-26 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structural basis of ketamine action on human NMDA receptors.
Nature, 596, 2021
|
|
2A5S
 
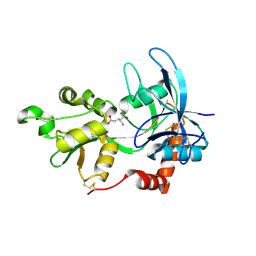 | | Crystal Structure Of The NR2A Ligand Binding Core In Complex With Glutamate | | Descriptor: | GLUTAMIC ACID, N-methyl-D-aspartate receptor NMDAR2A subunit | | Authors: | Furukawa, H, Singh, S.K, Mancusso, R, Gouaux, E. | | Deposit date: | 2005-06-30 | | Release date: | 2005-11-15 | | Last modified: | 2017-07-26 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Subunit arrangement and function in NMDA receptors
Nature, 438, 2005
|
|
2A5T
 
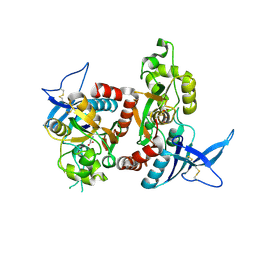 | | Crystal Structure Of The NR1/NR2A ligand-binding cores complex | | Descriptor: | GLUTAMIC ACID, GLYCINE, N-methyl-D-aspartate receptor NMDAR1-4a subunit, ... | | Authors: | Furukawa, H, Singh, S.K, Mancusso, R, Gouaux, E. | | Deposit date: | 2005-06-30 | | Release date: | 2005-11-15 | | Last modified: | 2017-07-26 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Subunit arrangement and function in NMDA receptors
Nature, 438, 2005
|
|
2AL5
 
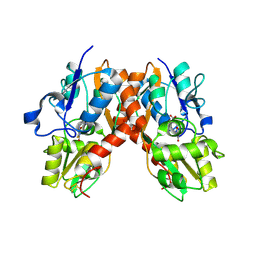 | | Crystal structure of the GluR2 ligand binding core (S1S2J) in complex with fluoro-willardiine and aniracetam | | Descriptor: | 1-(4-METHOXYBENZOYL)-2-PYRROLIDINONE, 2-AMINO-3-(5-FLUORO-2,4-DIOXO-3,4-DIHYDRO-2H-PYRIMIDIN-1-YL)-PROPIONIC ACID, Glutamate receptor 2 | | Authors: | Jin, R, Clark, S, Weeks, A.M, Dudman, J.T, Gouaux, E, Partin, K.M. | | Deposit date: | 2005-08-04 | | Release date: | 2005-10-25 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Mechanism of positive allosteric modulators acting on AMPA receptors.
J.Neurosci., 25, 2005
|
|
2ANJ
 
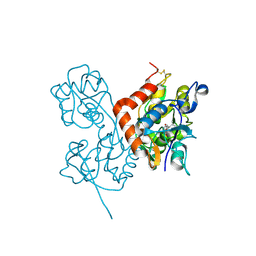 | | Crystal Structure of the Glur2 Ligand Binding Core (S1S2J-Y450W) Mutant in Complex With the Partial Agonist Kainic Acid at 2.1 A Resolution | | Descriptor: | 3-(CARBOXYMETHYL)-4-ISOPROPENYLPROLINE, Glutamate receptor 2 | | Authors: | Holm, M.M, Naur, P, Vestergaard, B, Geballe, M.T, Gajhede, M, Kastrup, J.S, Traynelis, S.F, Egebjerg, J. | | Deposit date: | 2005-08-11 | | Release date: | 2005-08-30 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | A Binding Site Tyrosine Shapes Desensitization Kinetics and Agonist Potency at GluR2: a mutagenic, kinetic, and crystallographic study
J.Biol.Chem., 280, 2005
|
|
2AL4
 
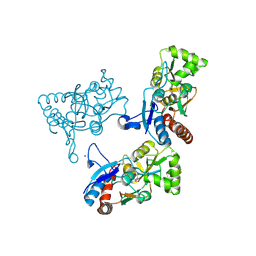 | | CRYSTAL STRUCTURE OF THE GLUR2 LIGAND BINDING CORE (S1S2J) IN COMPLEX WITH quisqualate and CX614. | | Descriptor: | (S)-2-AMINO-3-(3,5-DIOXO-[1,2,4]OXADIAZOLIDIN-2-YL)-PROPIONIC ACID, 2,3,6A,7,8,9-HEXAHYDRO-11H-[1,4]DIOXINO[2,3-G]PYRROLO[2,1-B][1,3]BENZOXAZIN-11-ONE, Glutamate receptor 2, ... | | Authors: | Jin, R, Clark, S, Weeks, A.M, Dudman, J.T, Gouaux, E, Partin, K.M. | | Deposit date: | 2005-08-04 | | Release date: | 2005-10-25 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Mechanism of positive allosteric modulators acting on AMPA receptors.
J.Neurosci., 25, 2005
|
|
2AIX
 
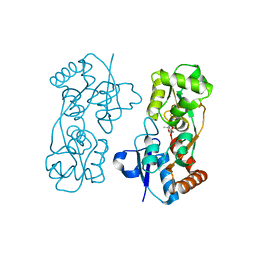 | |
6FQH
 
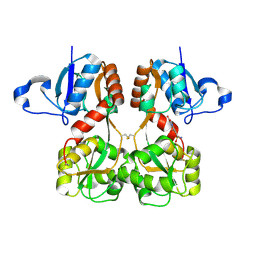 | | GluA2(flop) S729C ligand binding core dimer bound to NBQX at 1.76 Angstrom resolution | | Descriptor: | 6-nitro-2,3-bis(oxidanylidene)-1,4-dihydrobenzo[f]quinoxaline-7-sulfonamide, Glutamate receptor 2,Glutamate receptor 2 | | Authors: | Coombs, I.D, Soto, D, Gold, M.G, Farrant, M.F, Cull-Candy, S.G. | | Deposit date: | 2018-02-14 | | Release date: | 2019-03-13 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.75940013 Å) | | Cite: | Homomeric GluA2(R) AMPA receptors can conduct when desensitized.
Nat Commun, 10, 2019
|
|
6GL4
 
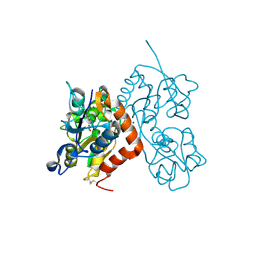 | | Structure of GluA2o ligand-binding domain (S1S2J) in complex with glutamate and sodium bromide at 1.95 A resolution | | Descriptor: | ACETATE ION, BROMIDE ION, GLUTAMIC ACID, ... | | Authors: | Venskutonyte, R, Frydenvang, K, Kastrup, J.S. | | Deposit date: | 2018-05-22 | | Release date: | 2019-05-15 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.948 Å) | | Cite: | Nanoscale Mobility of the Apo State and TARP Stoichiometry Dictate the Gating Behavior of Alternatively Spliced AMPA Receptors.
Neuron, 102, 2019
|
|
6GIV
 
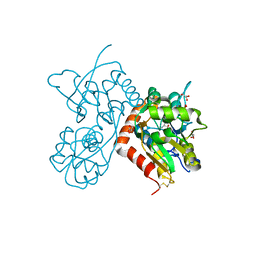 | | Structure of GluA2-N775S ligand-binding domain (S1S2J) in complex with glutamate and Rubidium Bromide at 1.75 A resolution | | Descriptor: | BROMIDE ION, GLUTAMIC ACID, GLYCEROL, ... | | Authors: | Venskutonyte, R, Frydenvang, K, Kastrup, J.S. | | Deposit date: | 2018-05-15 | | Release date: | 2019-05-15 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Nanoscale Mobility of the Apo State and TARP Stoichiometry Dictate the Gating Behavior of Alternatively Spliced AMPA Receptors.
Neuron, 102, 2019
|
|
6HCC
 
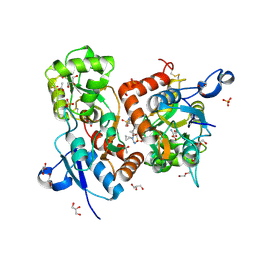 | | STRUCTURE OF GLUA2 LIGAND-BINDING DOMAIN (S1S2J-N775S) IN COMPLEX WITH GLUTAMATE AND TDPAM02 AT 1.6 A RESOLUTION. | | Descriptor: | 6,6'-(ETHANE-1,2-DIYL)BIS(4-CYCLOPROPYL-3,4-DIHYDRO-2H-1,2,4-BENZOTHIADIAZINE 1,1-DIOXIDE), ACETATE ION, CHLORIDE ION, ... | | Authors: | Laulumaa, S, Hansen, K.V, Frydenvang, K, Kastrup, J.S. | | Deposit date: | 2018-08-14 | | Release date: | 2019-04-03 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.617 Å) | | Cite: | Crystal Structures of Potent Dimeric Positive Allosteric Modulators at the Ligand-Binding Domain of the GluA2 Receptor.
Acs Med.Chem.Lett., 10, 2019
|
|
6HC9
 
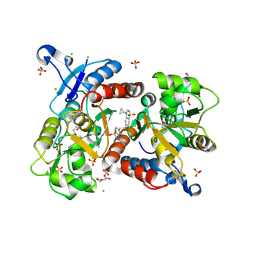 | | STRUCTURE OF GLUA2 LIGAND-BINDING DOMAIN (S1S2J-L504Y-N775S) IN COMPLEX WITH GLUTAMATE AND TDPAM02 AT 2.4 A RESOLUTION. | | Descriptor: | 6,6'-(ETHANE-1,2-DIYL)BIS(4-CYCLOPROPYL-3,4-DIHYDRO-2H-1,2,4-BENZOTHIADIAZINE 1,1-DIOXIDE), CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Laulumaa, S, Hansen, K.V, Frydenvang, K, Kastrup, J.S. | | Deposit date: | 2018-08-14 | | Release date: | 2019-04-03 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal Structures of Potent Dimeric Positive Allosteric Modulators at the Ligand-Binding Domain of the GluA2 Receptor.
Acs Med.Chem.Lett., 10, 2019
|
|
6HCA
 
 | | STRUCTURE OF GLUA2 LIGAND-BINDING DOMAIN (S1S1J) IN COMPLEX WITH POSITIVE ALLOSTERIC MODULATOR TDPAM02 AT 1.8 A RESOLUTION | | Descriptor: | 6,6'-(ETHANE-1,2-DIYL)BIS(4-CYCLOPROPYL-3,4-DIHYDRO-2H-1,2,4-BENZOTHIADIAZINE 1,1-DIOXIDE), CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Laulumaa, S, Hansen, K.V, Frydenvang, K, Kastrup, J.S. | | Deposit date: | 2018-08-14 | | Release date: | 2019-04-03 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.882 Å) | | Cite: | Crystal Structures of Potent Dimeric Positive Allosteric Modulators at the Ligand-Binding Domain of the GluA2 Receptor.
Acs Med.Chem.Lett., 10, 2019
|
|
6HCB
 
 | | STRUCTURE OF GLUA2 LIGAND-BINDING DOMAIN (S1S2J-N775S) IN COMPLEX WITH GLUTAMATE AND TDPAM01 AT 1.9 A RESOLUTION. | | Descriptor: | 6,6'-(Ethane-1,2-diyl)bis(4-methyl-3,4-dihydro-2H-1,2,4-benzothiadiazine 1,1-dioxide), CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Laulumaa, S, Masternak, M, Frydenvang, K, Kastrup, J.S. | | Deposit date: | 2018-08-14 | | Release date: | 2019-04-03 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal Structures of Potent Dimeric Positive Allosteric Modulators at the Ligand-Binding Domain of the GluA2 Receptor.
Acs Med.Chem.Lett., 10, 2019
|
|
6HCH
 
 | | STRUCTURE OF GLUA2 LIGAND-BINDING DOMAIN (S1S2J-L504Y-N775S) IN COMPLEX WITH GLUTAMATE AND TDPAM01 AT 1.6 A RESOLUTION. | | Descriptor: | 6,6'-(Ethane-1,2-diyl)bis(4-methyl-3,4-dihydro-2H-1,2,4-benzothiadiazine 1,1-dioxide), ACETATE ION, CHLORIDE ION, ... | | Authors: | Laulumaa, S, Hansen, K.V, Frydenvang, K, Kastrup, J.S. | | Deposit date: | 2018-08-15 | | Release date: | 2019-04-03 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal Structures of Potent Dimeric Positive Allosteric Modulators at the Ligand-Binding Domain of the GluA2 Receptor.
Acs Med.Chem.Lett., 10, 2019
|
|
6IRG
 
 | | Structure of the human GluN1/GluN2A NMDA receptor in the glutamate/glycine-bound state at pH 6.3, Class II | | Descriptor: | Glutamate receptor ionotropic, NMDA 1, NMDA 2A | | Authors: | Zhang, J, Chang, S, Zhang, X, Zhu, S. | | Deposit date: | 2018-11-12 | | Release date: | 2019-01-16 | | Last modified: | 2019-06-05 | | Method: | ELECTRON MICROSCOPY (5.5 Å) | | Cite: | Structural Basis of the Proton Sensitivity of Human GluN1-GluN2A NMDA Receptors
Cell Rep, 25, 2018
|
|
6IRA
 
 | | Structure of the human GluN1/GluN2A NMDA receptor in the glutamate/glycine-bound state at pH 7.8 | | Descriptor: | Glutamate receptor ionotropic, NMDA 1, NMDA 2A | | Authors: | Zhang, J, Chang, S, Zhang, X, Zhu, S. | | Deposit date: | 2018-11-12 | | Release date: | 2019-01-16 | | Last modified: | 2019-06-05 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | Structural Basis of the Proton Sensitivity of Human GluN1-GluN2A NMDA Receptors
Cell Rep, 25, 2018
|
|
6IRF
 
 | | Structure of the human GluN1/GluN2A NMDA receptor in the glutamate/glycine-bound state at pH 6.3, Class I | | Descriptor: | Glutamate receptor ionotropic, NMDA 1, NMDA 2A | | Authors: | Zhang, J, Chang, S, Zhang, X, Zhu, S. | | Deposit date: | 2018-11-12 | | Release date: | 2019-01-16 | | Last modified: | 2019-06-05 | | Method: | ELECTRON MICROSCOPY (5.1 Å) | | Cite: | Structural Basis of the Proton Sensitivity of Human GluN1-GluN2A NMDA Receptors
Cell Rep, 25, 2018
|
|
6IRH
 
 | | Structure of the human GluN1/GluN2A NMDA receptor in the glutamate/glycine-bound state at pH 6.3, Class III | | Descriptor: | Glutamate receptor ionotropic, NMDA 1, NMDA 2A | | Authors: | Zhang, J, Chang, S, Zhang, X, Zhu, S. | | Deposit date: | 2018-11-12 | | Release date: | 2019-01-16 | | Last modified: | 2019-06-05 | | Method: | ELECTRON MICROSCOPY (7.8 Å) | | Cite: | Structural Basis of the Proton Sensitivity of Human GluN1-GluN2A NMDA Receptors
Cell Rep, 25, 2018
|
|
6JFY
 
 | | GluK3 receptor trapped in Desensitized state | | Descriptor: | Glutamate receptor ionotropic, kainate 3 | | Authors: | Kumari, J, Kumar, J. | | Deposit date: | 2019-02-13 | | Release date: | 2019-07-24 | | Last modified: | 2019-11-06 | | Method: | ELECTRON MICROSCOPY (7.4 Å) | | Cite: | Structural and Functional Insights into GluK3-kainate Receptor Desensitization and Recovery.
Sci Rep, 9, 2019
|
|
6JFZ
 
 | | GluK3 receptor complex with UBP310 | | Descriptor: | Glutamate receptor ionotropic, kainate 3 | | Authors: | Kumari, J, Kumar, J. | | Deposit date: | 2019-02-13 | | Release date: | 2019-07-24 | | Last modified: | 2019-11-06 | | Method: | ELECTRON MICROSCOPY (7.6 Å) | | Cite: | Structural and Functional Insights into GluK3-kainate Receptor Desensitization and Recovery.
Sci Rep, 9, 2019
|
|
