7SAA
 
 | |
7SAD
 
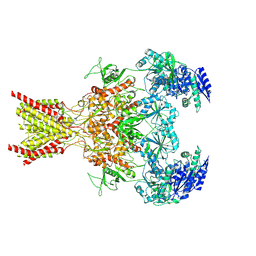 | | Memantine-bound GluN1a-GluN2B NMDA receptors | | 分子名称: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Glutamate receptor ionotropic, ... | | 著者 | Chou, T.-H, Furukawa, H. | | 登録日 | 2021-09-22 | | 公開日 | 2022-07-20 | | 実験手法 | ELECTRON MICROSCOPY (3.96 Å) | | 主引用文献 | Structural insights into binding of therapeutic channel blockers in NMDA receptors.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7SAB
 
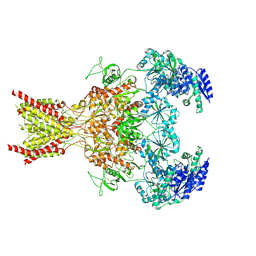 | | Phencyclidine-bound GluN1a-GluN2B NMDA receptors | | 分子名称: | 1-(PHENYL-1-CYCLOHEXYL)PIPERIDINE, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | 著者 | Chou, T.-H, Furukawa, H. | | 登録日 | 2021-09-22 | | 公開日 | 2022-07-20 | | 実験手法 | ELECTRON MICROSCOPY (4.3 Å) | | 主引用文献 | Structural insights into binding of therapeutic channel blockers in NMDA receptors.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7TER
 
 | |
7TEE
 
 | |
7TEB
 
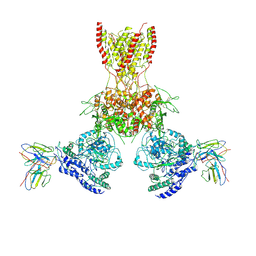 | |
7TES
 
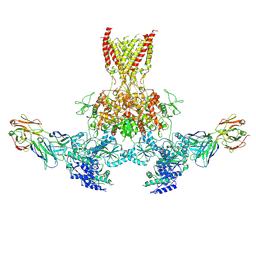 | |
7TET
 
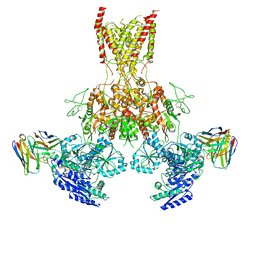 | |
7TE9
 
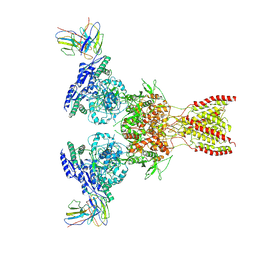 | |
7TEQ
 
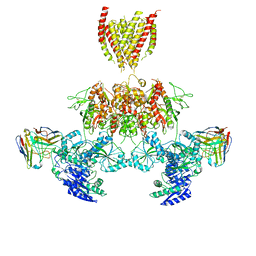 | |
3BFT
 
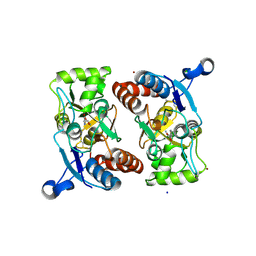 | | Structure of the ligand-binding core of GluR2 in complex with the agonist (S)-TDPA at 2.25 A resolution | | 分子名称: | (2S)-2-amino-3-(4-hydroxy-1,2,5-thiadiazol-3-yl)propanoic acid, CACODYLATE ION, CHLORIDE ION, ... | | 著者 | Beich-Frandsen, M, Mirza, O, Vestergaard, B, Gajhede, M, Kastrup, J.S. | | 登録日 | 2007-11-23 | | 公開日 | 2008-10-28 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (2.27 Å) | | 主引用文献 | Structures of the ligand-binding core of iGluR2 in complex with the agonists (R)- and (S)-2-amino-3-(4-hydroxy-1,2,5-thiadiazol-3-yl)propionic acid explain their unusual equipotency.
J.Med.Chem., 51, 2008
|
|
3B6T
 
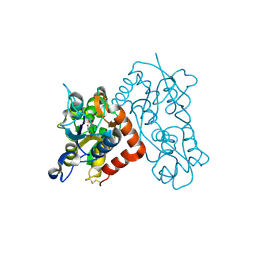 | | Crystal Structure of the GLUR2 Ligand Binding Core (S1S2J) T686A Mutant in Complex with Quisqualate at 2.1 Resolution | | 分子名称: | (S)-2-AMINO-3-(3,5-DIOXO-[1,2,4]OXADIAZOLIDIN-2-YL)-PROPIONIC ACID, Glutamate receptor 2, SULFATE ION | | 著者 | Cho, Y, Lolis, E, Howe, J.R. | | 登録日 | 2007-10-29 | | 公開日 | 2008-02-05 | | 最終更新日 | 2021-10-20 | | 実験手法 | X-RAY DIFFRACTION (2.1 Å) | | 主引用文献 | Structural and single-channel results indicate that the rates of ligand binding domain closing and opening directly impact AMPA receptor gating.
J.Neurosci., 28, 2008
|
|
3B7D
 
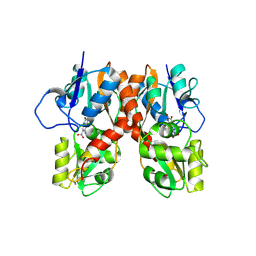 | |
3BBR
 
 | |
3B6Q
 
 | |
3B6W
 
 | |
3BFU
 
 | | Structure of the ligand-binding core of GluR2 in complex with the agonist (R)-TDPA at 1.95 A resolution | | 分子名称: | (2R)-2-amino-3-(4-hydroxy-1,2,5-thiadiazol-3-yl)propanoic acid, Glutamate receptor 2 | | 著者 | Beich-Frandsen, M, Mirza, O, Vestergaard, B, Gajhede, M, Kastrup, J.S. | | 登録日 | 2007-11-23 | | 公開日 | 2008-10-14 | | 最終更新日 | 2023-11-01 | | 実験手法 | X-RAY DIFFRACTION (1.95 Å) | | 主引用文献 | Structures of the ligand-binding core of iGluR2 in complex with the agonists (R)- and (S)-2-amino-3-(4-hydroxy-1,2,5-thiadiazol-3-yl)propionic acid explain their unusual equipotency.
J.Med.Chem., 51, 2008
|
|
3C33
 
 | |
3C36
 
 | |
3C34
 
 | |
3C35
 
 | |
3C32
 
 | |
3BKI
 
 | | Crystal Structure of the GluR2 ligand binding core (S1S2J) in complex with FQX at 1.87 Angstroms | | 分子名称: | Glutamate receptor 2, [1,2,5]oxadiazolo[3,4-g]quinoxaline-6,7(5H,8H)-dione 1-oxide | | 著者 | Cruz, L, Estebanez-Perpina, E, Pfaff, S, Borngraeber, S, Bao, N, Fletterick, R, England, P. | | 登録日 | 2007-12-06 | | 公開日 | 2008-09-16 | | 最終更新日 | 2019-09-04 | | 実験手法 | X-RAY DIFFRACTION (1.87 Å) | | 主引用文献 | 6-Azido-7-nitro-1,4-dihydroquinoxaline-2,3-dione (ANQX) forms an irreversible bond to the active site of the GluR2 AMPA receptor.
J.Med.Chem., 51, 2008
|
|
3C31
 
 | |
4IO4
 
 | | Crystal Structure of the AvGluR1 ligand binding domain complex with serine at 1.94 Angstrom resolution | | 分子名称: | AvGluR1 ligand binding domain, CHLORIDE ION, GLYCEROL, ... | | 著者 | Lomash, S, Chittori, S, Mayer, M.L. | | 登録日 | 2013-01-07 | | 公開日 | 2013-02-20 | | 最終更新日 | 2023-09-20 | | 実験手法 | X-RAY DIFFRACTION (1.941 Å) | | 主引用文献 | Anions Mediate Ligand Binding in Adineta vaga Glutamate Receptor Ion Channels.
Structure, 21, 2013
|
|
