6TEJ
 
 | | Structure of apo IrtAB devoid SID in complex with sybody Syb_NL5 | | Descriptor: | DECYL-BETA-D-MALTOPYRANOSIDE, Drug ABC transporter ATP-binding protein, NICKEL (II) ION, ... | | Authors: | Gonda, I, Arnold, F.M, Hutter, C.A.J, Weber, M.S, Seeger, M.A, Hurlimann, L.M. | | Deposit date: | 2019-11-12 | | Release date: | 2020-04-01 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | The ABC exporter IrtAB imports and reduces mycobacterial siderophores.
Nature, 580, 2020
|
|
6TQE
 
 | | The structure of ABC transporter Rv1819c without addition of substrate | | Descriptor: | ABC transporter ATP-binding protein/permease, ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION | | Authors: | Rempel, S, Gati, C, Slotboom, D.J, Guskov, A. | | Deposit date: | 2019-12-16 | | Release date: | 2020-04-01 | | Last modified: | 2021-02-10 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | A mycobacterial ABC transporter mediates the uptake of hydrophilic compounds.
Nature, 580, 2020
|
|
6TMF
 
 | | Structure of an archaeal ABCE1-bound ribosomal post-splitting complex | | Descriptor: | 16S ribosomal RNA, 30S ribosomal protein S10, 30S ribosomal protein S11, ... | | Authors: | Kratzat, H, Becker, T, Tampe, R, Beckmann, R. | | Deposit date: | 2019-12-04 | | Release date: | 2020-02-12 | | Last modified: | 2020-05-13 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Molecular analysis of the ribosome recycling factor ABCE1 bound to the 30S post-splitting complex.
Embo J., 39, 2020
|
|
6UJT
 
 | | P-glycoprotein mutant-Y303A and C952A-with BDE100 | | Descriptor: | 2,4-dibromophenyl 2,4,6-tribromophenyl ether, ATP-dependent translocase ABCB1 | | Authors: | Aller, S.G, Le, C.A. | | Deposit date: | 2019-10-03 | | Release date: | 2020-05-27 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (4.17 Å) | | Cite: | Structural definition of polyspecific compensatory ligand recognition by P-glycoprotein.
Iucrj, 7, 2020
|
|
6UJR
 
 | | P-glycoprotein mutant-F724A and C952A-with BDE100 | | Descriptor: | 2,4-dibromophenyl 2,4,6-tribromophenyl ether, ATP-dependent translocase ABCB1 | | Authors: | Aller, S.G, Le, C.A. | | Deposit date: | 2019-10-03 | | Release date: | 2020-05-27 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (4.1 Å) | | Cite: | Structural definition of polyspecific compensatory ligand recognition by P-glycoprotein.
Iucrj, 7, 2020
|
|
6UJW
 
 | | P-glycoprotein mutant-Y306A and C952A-with BDE100 | | Descriptor: | 2,4-dibromophenyl 2,4,6-tribromophenyl ether, ATP-dependent translocase ABCB1 | | Authors: | Aller, S.G, Le, C.A. | | Deposit date: | 2019-10-03 | | Release date: | 2020-05-27 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (4.15 Å) | | Cite: | Structural definition of polyspecific compensatory ligand recognition by P-glycoprotein.
Iucrj, 7, 2020
|
|
6UJP
 
 | | P-glycoprotein mutant-F979A and C952A-with BDE100 | | Descriptor: | 2,4-dibromophenyl 2,4,6-tribromophenyl ether, ATP-dependent translocase ABCB1 | | Authors: | Aller, S.G, Le, C.A. | | Deposit date: | 2019-10-03 | | Release date: | 2020-05-27 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.98 Å) | | Cite: | Structural definition of polyspecific compensatory ligand recognition by P-glycoprotein.
Iucrj, 7, 2020
|
|
6UJS
 
 | | P-glycoprotein mutant-F728A and C952A-with BDE100 | | Descriptor: | 2,4-dibromophenyl 2,4,6-tribromophenyl ether, ATP-dependent translocase ABCB1 | | Authors: | Aller, S.G, Le, C.A. | | Deposit date: | 2019-10-03 | | Release date: | 2020-05-27 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (4.17 Å) | | Cite: | Structural definition of polyspecific compensatory ligand recognition by P-glycoprotein.
Iucrj, 7, 2020
|
|
6UJN
 
 | | P-glycoprotein mutant-C952A-with BDE100 | | Descriptor: | 2,4-dibromophenyl 2,4,6-tribromophenyl ether, ATP-dependent translocase ABCB1 | | Authors: | Aller, S.G, Le, C.A. | | Deposit date: | 2019-10-03 | | Release date: | 2020-05-27 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.98 Å) | | Cite: | Structural definition of polyspecific compensatory ligand recognition by P-glycoprotein.
Iucrj, 7, 2020
|
|
6V9Z
 
 | |
6UY0
 
 | |
6UZL
 
 | | Cryo-EM structure of nucleotide-free MsbA reconstituted into peptidiscs, conformation 2 | | Descriptor: | Lipid A export ATP-binding/permease protein MsbA | | Authors: | Angiulli, G, Walz, T, Dhupar, H.S, Suzuki, H, Wason, I.S, Duong Van Hoa, F. | | Deposit date: | 2019-11-15 | | Release date: | 2020-03-04 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (4.4 Å) | | Cite: | New approach for membrane protein reconstitution into peptidiscs and basis for their adaptability to different proteins.
Elife, 9, 2020
|
|
6UZ2
 
 | | Cryo-EM structure of nucleotide-free MsbA reconstituted into peptidiscs, conformation 1 | | Descriptor: | Lipid A export ATP-binding/permease protein MsbA | | Authors: | Angiulli, G, Walz, T, Dhupar, H.S, Suzuki, H, Wason, I.S, Duong Van Hoa, F. | | Deposit date: | 2019-11-14 | | Release date: | 2020-03-04 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | New approach for membrane protein reconstitution into peptidiscs and basis for their adaptability to different proteins.
Elife, 9, 2020
|
|
6VQU
 
 | | Structure of a bacterial Atm1-family ABC exporter | | Descriptor: | ATM1-type heavy metal exporter | | Authors: | Fan, C, Rees, D.C. | | Deposit date: | 2020-02-05 | | Release date: | 2020-07-29 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.88 Å) | | Cite: | A structural framework for unidirectional transport by a bacterial ABC exporter.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
6VXF
 
 | | Structure of apo-closed ABCG2 | | Descriptor: | Broad substrate specificity ATP-binding cassette transporter ABCG2 | | Authors: | Orlando, B.J, Maofu, L. | | Deposit date: | 2020-02-21 | | Release date: | 2020-05-13 | | Last modified: | 2020-05-20 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | ABCG2 transports anticancer drugs via a closed-to-open switch.
Nat Commun, 11, 2020
|
|
6VQT
 
 | |
6VXJ
 
 | | Structure of ABCG2 bound to SN38 | | Descriptor: | 7-ethyl-10-hydroxycamptothecin, Broad substrate specificity ATP-binding cassette transporter ABCG2, CHOLESTEROL | | Authors: | Orlando, B.J, Liao, M. | | Deposit date: | 2020-02-22 | | Release date: | 2020-05-13 | | Last modified: | 2020-05-20 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | ABCG2 transports anticancer drugs via a closed-to-open switch.
Nat Commun, 11, 2020
|
|
6VXH
 
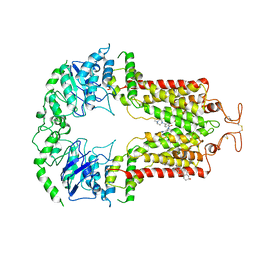 | | Structure of ABCG2 bound to imatinib | | Descriptor: | 4-(4-METHYL-PIPERAZIN-1-YLMETHYL)-N-[4-METHYL-3-(4-PYRIDIN-3-YL-PYRIMIDIN-2-YLAMINO)-PHENYL]-BENZAMIDE, Broad substrate specificity ATP-binding cassette transporter ABCG2, CHOLESTEROL | | Authors: | Orlando, B.J, Liao, M. | | Deposit date: | 2020-02-21 | | Release date: | 2020-05-13 | | Last modified: | 2020-05-20 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | ABCG2 transports anticancer drugs via a closed-to-open switch.
Nat Commun, 11, 2020
|
|
6VXI
 
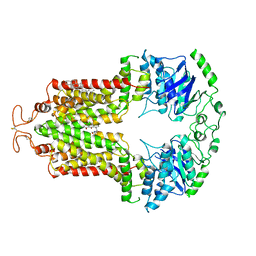 | | Structure of ABCG2 bound to mitoxantrone | | Descriptor: | 1,4-DIHYDROXY-5,8-BIS({2-[(2-HYDROXYETHYL)AMINO]ETHYL}AMINO)-9,10-ANTHRACENEDIONE, Broad substrate specificity ATP-binding cassette transporter ABCG2, CHOLESTEROL | | Authors: | Orlando, B.J, Liao, M. | | Deposit date: | 2020-02-21 | | Release date: | 2020-05-13 | | Last modified: | 2020-05-20 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | ABCG2 transports anticancer drugs via a closed-to-open switch.
Nat Commun, 11, 2020
|
|
5UAK
 
 | |
5TV4
 
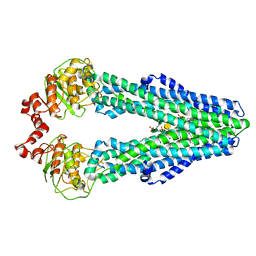 | | 3D cryo-EM reconstruction of nucleotide-free MsbA in lipid nanodisc | | Descriptor: | 3-HYDROXY-TETRADECANOIC ACID, L-glycero-alpha-D-manno-heptopyranose-(1-7)-L-glycero-alpha-D-manno-heptopyranose-(1-3)-L-glycero-alpha-D-manno-heptopyranose-(1-5)-[3-deoxy-alpha-D-manno-oct-2-ulopyranosonic acid-(2-4)]3-deoxy-alpha-D-manno-oct-2-ulopyranosonic acid-(2-6)-2-amino-2-deoxy-alpha-D-glucopyranose-(1-6)-2-amino-2-deoxy-alpha-D-glucopyranose, LAURIC ACID, ... | | Authors: | Mi, W, Walz, T, Liao, M. | | Deposit date: | 2016-11-08 | | Release date: | 2017-09-20 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Structural basis of MsbA-mediated lipopolysaccharide transport.
Nature, 549, 2017
|
|
5TTP
 
 | |
5U1D
 
 | |
5TWV
 
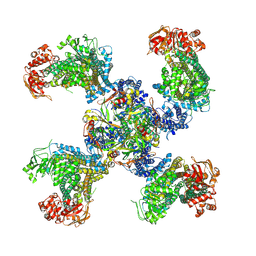 | | Cryo-EM structure of the pancreatic ATP-sensitive K+ channel SUR1/Kir6.2 in the presence of ATP and glibenclamide | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, ATP-binding cassette sub-family C member 8, ATP-sensitive inward rectifier potassium channel 11 | | Authors: | Martin, G.M, Yoshioka, C, Chen, J.Z, Shyng, S.L. | | Deposit date: | 2016-11-14 | | Release date: | 2017-01-25 | | Last modified: | 2019-12-25 | | Method: | ELECTRON MICROSCOPY (6.3 Å) | | Cite: | Cryo-EM structure of the ATP-sensitive potassium channel illuminates mechanisms of assembly and gating.
Elife, 6, 2017
|
|
5UAR
 
 | |
